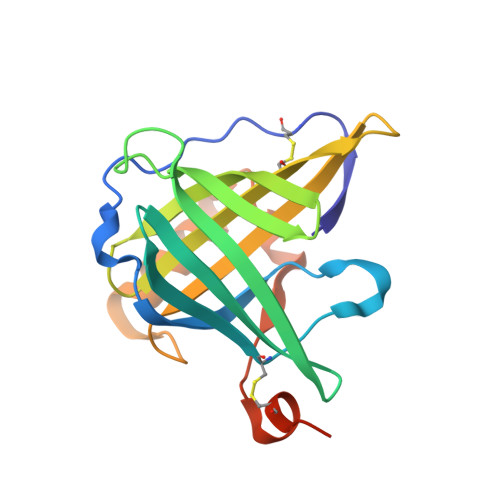
Crystallographic analysis of an "anticalin" with tailored specificity for fluorescein reveals high structural plasticity of the lipocalin loop region.
Korndorfer, I.P., Beste, G., Skerra, A.(2003) Proteins 53: 121-129
- PubMed: 12945055
- DOI: https://doi.org/10.1002/prot.10497
- Primary Citation of Related Structures:
1N0S - PubMed Abstract:
The artificial lipocalin FluA with novel specificity toward fluorescein was derived via combinatorial engineering from the bilin-binding protein, BBP by exchange of 16 amino acids in the ligand pocket. Here, we describe the crystal structure of FluA at 2.0 A resolution in the space group P2(1) with two protein-ligand complexes in the asymmetric unit. In both molecules, the characteristic beta-barrel architecture with the attached alpha-helix is well preserved. In contrast, the four loops at one end of the beta-barrel that form the entrance to the binding site exhibit large conformational deviations from the wild-type protein, which can be attributed to the sidechain replacements. Specificity for the new ligand is furnished by hydrophobic packing, charged sidechain environment, and hydrogen bonds with its hydroxyl groups. Unexpectedly, fluorescein is bound in a much deeper cavity than biliverdin IX(gamma) in the natural lipocalin. Triggered by the substituted residues, unmutated sidechains at the bottom of the binding site adopt conformations that are quite different from those observed in the BBP, illustrating that not only the loop region but also the hydrophobic interior of the beta-barrel can be reshaped for molecular recognition. Particularly, Trp 129 participates in a tight stacking interaction with the xanthenolone moiety, which may explain the ultrafast electron transfer that occurs on light excitation of the bound fluorescein. These structural findings support our concept of using lipocalins as a scaffold for the engineering of so-called "anticalins" directed against prescribed targets as an alternative to recombinant antibody fragments.
- Lehrstuhl für Biologische Chemie, Technische Universität München, Freising-Weihenstephan, Germany.
Organizational Affiliation: