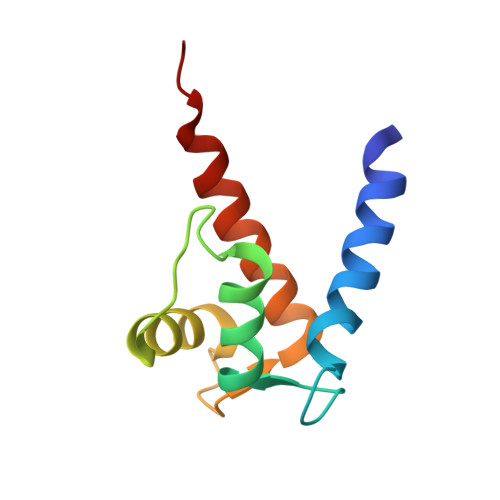
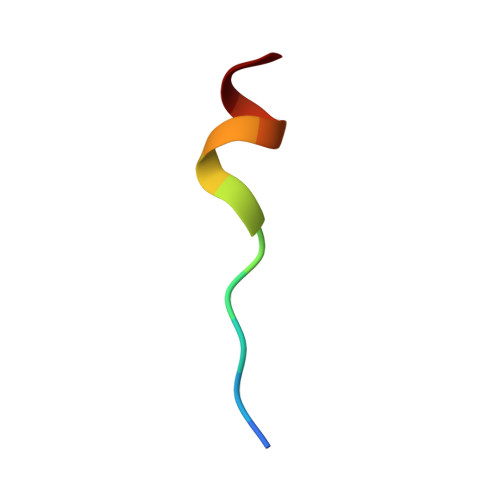Solution NMR structure of S100B bound to the high-affinity target peptide TRTK-12
Inman, K.G., Yang, R., Rustandi, R.R., Miller, K.E., Baldisseri, D.M., Weber, D.J.(2002) J Mol Biology 324: 1003-1014
- PubMed: 12470955
- DOI: https://doi.org/10.1016/s0022-2836(02)01152-x
- Primary Citation of Related Structures:
1MWN - PubMed Abstract:
The solution NMR structure is reported for Ca(2+)-loaded S100B bound to a 12-residue peptide, TRTK-12, from the actin capping protein CapZ (alpha1 or alpha2 subunit, residues 265-276: TRTKIDWNKILS). This peptide was discovered by Dimlich and co-workers by screening a bacteriophage random peptide display library, and it matches exactly the consensus S100B binding sequence ((K/R)(L/I)XWXXIL). As with other S100B target proteins, a calcium-dependent conformational change in S100B is required for TRTK-12 binding. The TRTK-12 peptide is an amphipathic helix (residues W7 to S12) in the S100B-TRTK complex, and helix 4 of S100B is extended by three or four residues upon peptide binding. However, helical TRTK-12 in the S100B-peptide complex is uniquely oriented when compared to the three-dimensional structures of other S100-peptide complexes. The three-dimensional structure of the S100B-TRTK peptide complex illustrates that residues in the S100B binding consensus sequence (K4, I5, W7, I10, L11) are all involved in the S100B-peptide interface, which can explain its orientation in the S100B binding pocket and its relatively high binding affinity. A comparison of the S100B-TRTK peptide structure to the structures of apo- and Ca(2+)-bound S100B illustrates that the binding site of TRTK-12 is buried in apo-S100B, but is exposed in Ca(2+)-bound S100B as necessary to bind the TRTK-12 peptide.
- Department of Biochemistry and Molecular Biology, School of Medicine, University of Maryland, 108 N. Greene St., Baltimore, MD 21201, USA.
Organizational Affiliation: