Determination of the three-dimensional structure of margatoxin by 1H, 13C, 15N triple-resonance nuclear magnetic resonance spectroscopy.
Johnson, B.A., Stevens, S.P., Williamson, J.M.(1994) Biochemistry 33: 15061-15070
- PubMed: 7999764
- DOI: https://doi.org/10.1021/bi00254a015
- Primary Citation of Related Structures:
1MTX - PubMed Abstract:
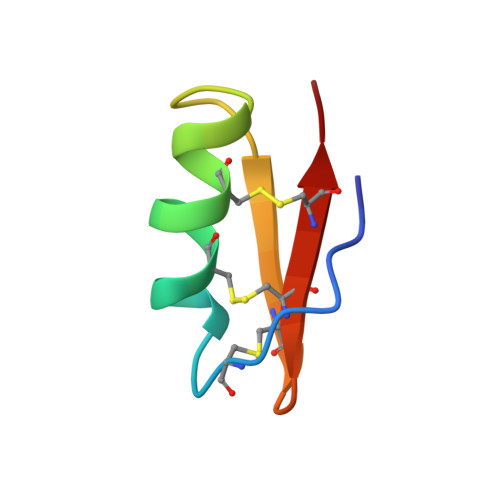
The solution structure of the 39-residue peptide margatoxin, a scorpion toxin that selectively blocks the voltage-gated potassium-channel Kv1.3, has been determined by NMR spectroscopy. The toxin was isotopically labeled with 13C and 15N and studied using two-dimensional homonuclear and three- and four-dimensional heteronuclear NMR spectroscopy. The final structure was determined using 501 constraints, comprising 422 NOE constraints, 60 dihedral angle constraints, 9 disulfide constraints, and 10 hydrogen bond constraints. Structures were initially determined with the program PEGASUS and subsequently refined with X-PLOR. The average rms deviation from a calculated average structure for the backbone atoms of residues 3-38 is 0.40 A. A helix is present from residues 11 to 20 and includes two proline residues at positions 15 and 16. A loop at residues 21-24 leads into a two-strand antiparallel sheet from residues 25 to 38 with a turn at residues 30-33. Residues 3-6 run adjacent to the 33-38 strand but do not form a canonical beta-strand. The two additional residues of margatoxin, relative to the related toxins charybdotoxin and iberiotoxin, insert in a manner that extends the beta-sheet by one residue. Otherwise, the global structure is very similar to that of these two other toxins. The longer sheet may have implications for channel selectivity.
- Department of Biophysical Chemistry, Merck Research Laboratories, Rahway, New Jersey 07065.
Organizational Affiliation: