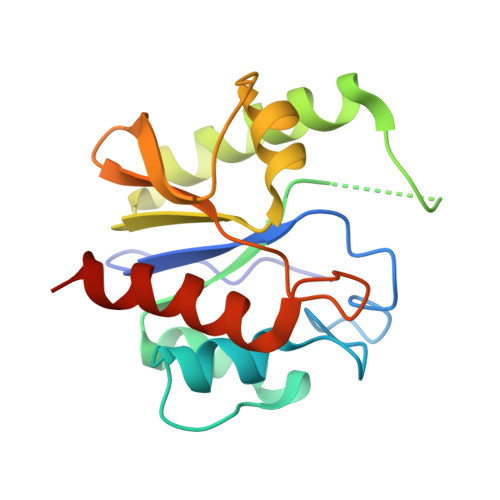
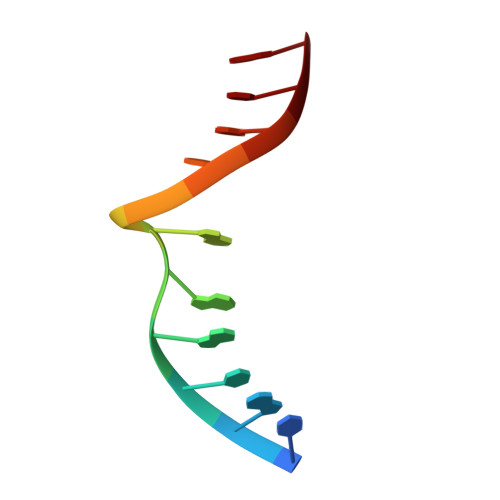
Structure of a DNA base-excision product resembling a cisplatin inter-strand adduct.
Barrett, T.E., Savva, R., Barlow, T., Brown, T., Jiricny, J., Pearl, L.H.(1998) Nat Struct Biol 5: 697-701
- PubMed: 9699633
- DOI: https://doi.org/10.1038/1394
- Primary Citation of Related Structures:
1MTL - PubMed Abstract:
Base-excision of a self-complementary oligonucleotide with central G:T mismatches by the G:T/U-specific mismatch DNA glycosylase (MUG), generates an unusual DNA structure which is remarkably similar in conformation to an interstrand DNA adduct of the anti-tumor drug cis-diamminedichloroplatinum. The abasic sugars generated by excision of the mismatched thymines are extruded from the double-helix, and the 'widowed' deoxyguanosines rotate so that their N7 and O6 groups protrude into the minor groove of the duplex and restack in an interleaved intercalative geometry, generating a kink in the helix axis.
- Department of Biochemistry and Molecular Biology, University College London, UK.
Organizational Affiliation: