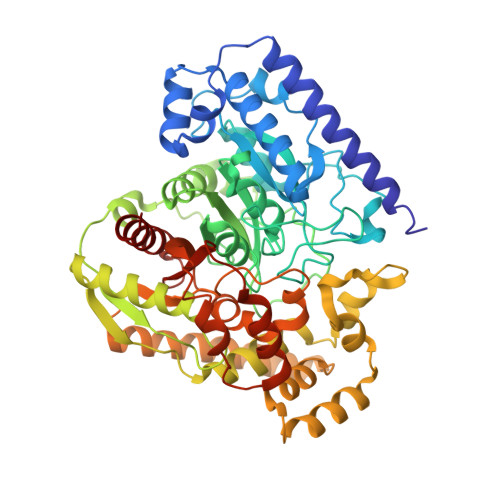
Structural Adaptations in a Membrane Enzyme That Terminates Endocannabinoid Signaling
Bracey, M.H., Hanson, M.A., Masuda, K.R., Stevens, R.C., Cravatt, B.F.(2002) Science 298: 1793-1796
- PubMed: 12459591
- DOI: https://doi.org/10.1126/science.1076535
- Primary Citation of Related Structures:
1MT5 - PubMed Abstract:
Cellular communication in the nervous system is mediated by chemical messengers that include amino acids, monoamines, peptide hormones, and lipids. An interesting question is how neurons regulate signals that are transmitted by membrane-embedded lipids. Here, we report the 2.8 angstrom crystal structure of the integral membrane protein fatty acid amide hydrolase (FAAH), an enzyme that degrades members of the endocannabinoid class of signaling lipids and terminates their activity. The structure of FAAH complexed with an arachidonyl inhibitor reveals how a set of discrete structural alterations allows this enzyme, in contrast to soluble hydrolases of the same family, to integrate into cell membranes and establish direct access to the bilayer from its active site.
- Department of Cell Biology, Skaggs Institute for Chemical Biology, Scripps Research Institute, 10550 North Torrey Pines Road, La Jolla, CA 92037, USA.
Organizational Affiliation: