Solution structure of a specific DNA complex of the Myb DNA-binding domain with cooperative recognition helices.
Ogata, K., Morikawa, S., Nakamura, H., Sekikawa, A., Inoue, T., Kanai, H., Sarai, A., Ishii, S., Nishimura, Y.(1994) Cell 79: 639-648
- PubMed: 7954830
- DOI: https://doi.org/10.1016/0092-8674(94)90549-5
- Primary Citation of Related Structures:
1MSE, 1MSF - PubMed Abstract:
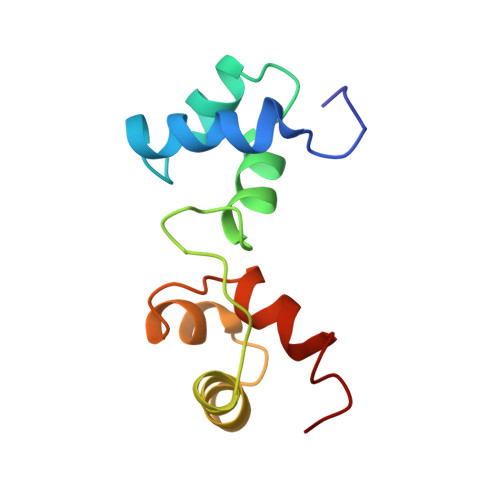
The DNA-binding region of Myb consists of three imperfect tandem repeats (R1, R2, and R3). We have determined the solution structure of a specific DNA complex of the minimum DNA-binding domain (R2R3) by heteronuclear multidimensional NMR. Both R2 and R3 contain three helices, and the third helix in each is found to be a recognition helix. R2 and R3 are closely packed in the major groove, so that the two recognition helices contact each other directly to bind to the specific base sequence, AACNG cooperatively; this is a significant arrangement of recognition helices. The three key base pairs in this sequence are specifically recognized by Asn-183 (R3), Lys-182 (R3), and Lys-128 (R2). In contrast, R1 has no specific interactions with DNA from our NMR study of the DNA complex of the full DNA-binding domain (R1R2R3).
- Graduate School of Integrated Science, Yokohama City University, Japan.
Organizational Affiliation: