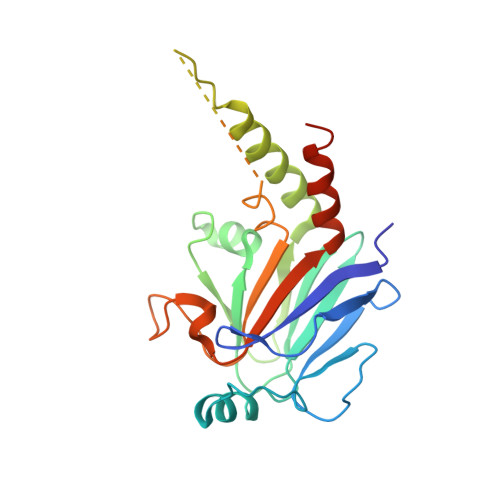
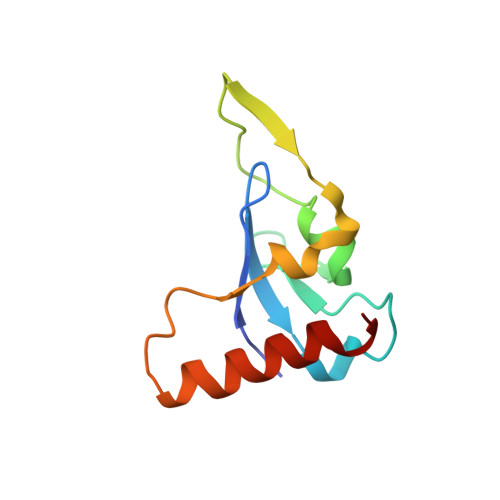
Structural Mechanism of Smad4 Recognition by the Nuclear Oncoprotein Ski: Insights on Ski-mediated Repression of TGF-beta Signaling
Wu, J.-W., Krawitz, A.R., Chai, J., Li, W., Zhang, F., Luo, K., Shi, Y.(2002) Cell 111: 357-367
- PubMed: 12419246
- DOI: https://doi.org/10.1016/s0092-8674(02)01006-1
- Primary Citation of Related Structures:
1MR1 - PubMed Abstract:
The Ski family of nuclear oncoproteins represses TGF-beta signaling through interactions with the Smad proteins. The crystal structure of the Smad4 binding domain of human c-Ski in complex with the MH2 domain of Smad4 reveals specific recognition of the Smad4 L3 loop region by a highly conserved interaction loop (I loop) from Ski. The Ski binding surface on Smad4 significantly overlaps with that required for binding of the R-Smads. Indeed, Ski disrupts the formation of a functional complex between the Co- and R-Smads, explaining how it could lead to repression of TGF-beta, activin, and BMP responses. Intriguingly, the structure of the Ski fragment, stabilized by a bound zinc atom, resembles the SAND domain, in which the corresponding I loop is responsible for DNA binding.
- Department of Molecular Biology, Lewis Thomas Laboratory, Princeton University, Princeton, NJ 08544, USA.
Organizational Affiliation: