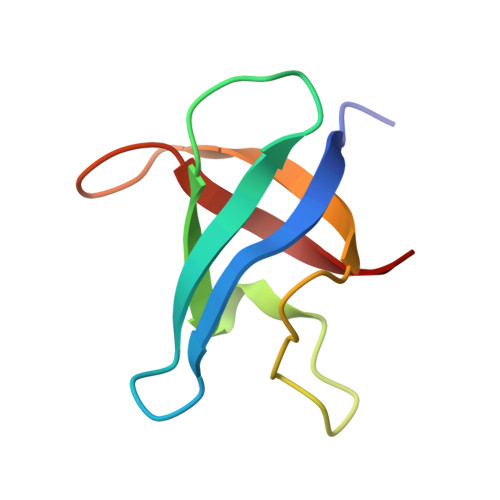
Crystal structure of CspA, the major cold shock protein of Escherichia coli.
Schindelin, H., Jiang, W., Inouye, M., Heinemann, U.(1994) Proc Natl Acad Sci U S A 91: 5119-5123
- PubMed: 8197194
- DOI: https://doi.org/10.1073/pnas.91.11.5119
- Primary Citation of Related Structures:
1MJC - PubMed Abstract:
The major cold shock protein of Escherichia coli, CspA, produced upon a rapid downshift in growth temperature, is involved in the transcriptional regulation of at least two genes. The protein shares high homology with the nucleic acid-binding domain of the Y-box factors, a family of eukaryotic proteins involved in transcriptional and translational regulation. The crystal structure of CspA has been determined at 2-A resolution and refined to R = 0.187. CspA is composed of five antiparallel beta-strands forming a closed five-stranded beta-barrel. The three-dimensional structure of CspA is similar to that of the major cold shock protein of Bacillus subtilis, CspB, which has recently been determined at 2.45-A resolution. However, in contrast to CspB, no dimer is formed in the crystal. The surface of CspA is characteristic for a protein interacting with single-stranded nucleic acids. Due to the high homology of the bacterial cold shock proteins with the Y-box factors, E. coli CspA and B. subtilis CspB define a structural framework for the common cold shock domain.
- Forschungsgruppe Kristallographie, Max-Delbrúck-Centrum für Molekulare Medizin, Berlin, Germany.
Organizational Affiliation: