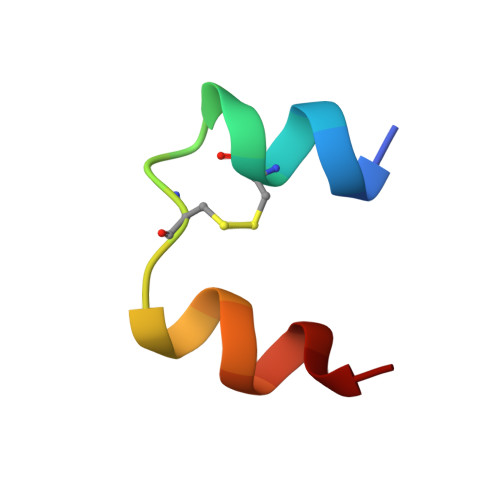
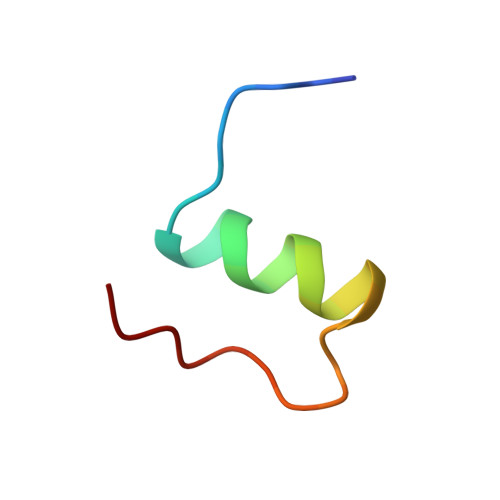
Solution structure of the superactive monomeric des-[Phe(B25)] human insulin mutant: elucidation of the structural basis for the monomerization of des-[Phe(B25)] insulin and the dimerization of native insulin.
Jorgensen, A.M., Olsen, H.B., Balschmidt, P., Led, J.J.(1996) J Mol Biology 257: 684-699
- PubMed: 8648633
- DOI: https://doi.org/10.1006/jmbi.1996.0194
- Primary Citation of Related Structures:
1MHJ - PubMed Abstract:
The three-dimensional solution structure of des-[Phe(B25)] human insulin has been determined by nuclear magnetic resonance spectroscopy and restrained molecular dynamics calculations. Thirty-five structures were calculated by distance geometry from 581 nuclear Overhauser enhancement-derived distance constraints, ten phi torsional angle restraints, the restraints from 16 helical hydrogen bonds, and three disulfide bridges. The distance geometry structures were optimized using simulated annealing and restrained energy minimization. The average root-mean-square (r.m.s.) deviation for the best 20 refined structures is 1.07 angstroms for the backbone and 1.92 angstroms for all atoms if the less well-defined N and C-terminal residues are excluded. The helical regions are more well defined, with r.m.s. deviations of 0.64 angstroms for the backbone and 1.51 angstroms for all atoms. It is found that the des-[Phe(B25)] insulin is a monomer under the applied conditions (4.6 to 4.7 mM, pH 3.0, 310 K), that the overall secondary and tertiary structures of the monomers in the 2Zn crystal hexamer of native insulin are preserved, and that the conformation-averaged NMR solution structure is close to the structure of molecule 1 in the hexamer. The structure reveals that the lost ability of des-[Phe(B25)] insulin to self-associate is caused by a conformational change of the C-terminal region of the B-chain, which results in an intra-molecular hydrophobic interaction between Pro(B28) and the hydrophobic region Leu(B11)-Leu(B15) of the B-chain alpha-helix. This interaction interferes with the inter-molecular hydrophobic interactions responsible for the dimerization of native insulin, depriving the mutant of the ability to dimerize. Further, the structure displays a series of features that may explain the high potency of the mutant on the basis of the current model for the insulin-receptor interaction. These features are: a change in conformation of the C-terminal region of the B-chain, the absence of strong hydrogen bonds between this region and the rest of the molecule, and a relatively easy accessibility to the Val(A3) residue.
- Department of Chemistry, University of Copenhagen, The H.C. Orsted Institute, Denmark.
Organizational Affiliation: