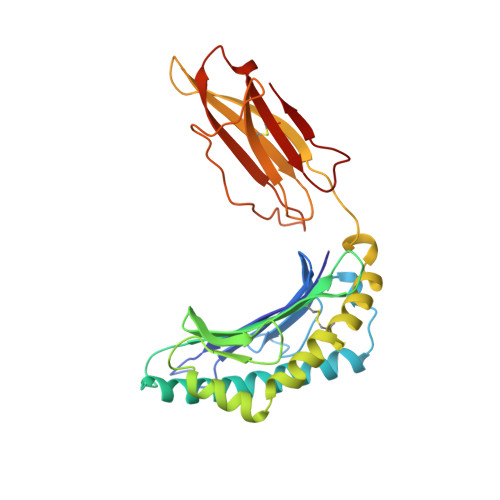
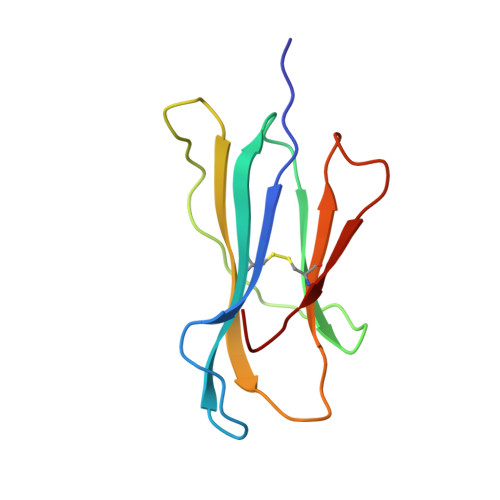
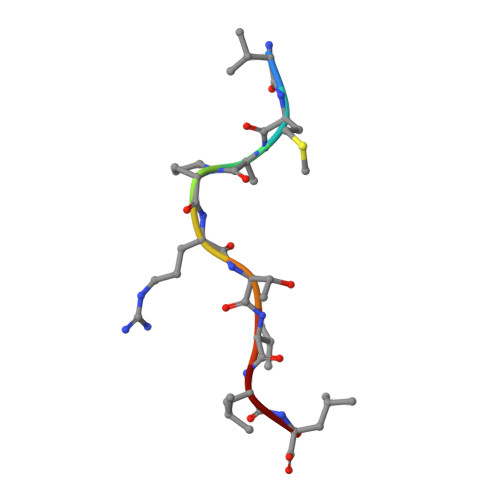
Structural features impose tight peptide binding specificity in the nonclassical MHC molecule HLA-E.
O'Callaghan, C.A., Tormo, J., Willcox, B.E., Braud, V.M., Jakobsen, B.K., Stuart, D.I., McMichael, A.J., Bell, J.I., Jones, E.Y.(1998) Mol Cell 1: 531-541
- PubMed: 9660937
- DOI: https://doi.org/10.1016/s1097-2765(00)80053-2
- Primary Citation of Related Structures:
1MHE - PubMed Abstract:
The crystal structure of the nonclassical human class lb MHC molecule HLA-E has been determined in complex with a prototypic ligand, the nonamer peptide (VMAPRTVLL), derived from the highly conserved residues 3-11 of the human MHC class la leader sequence. The mode of peptide binding retains some of the standard features observed in MHC class la complexes, but novel features imply that HLA-E has evolved to mediate specific binding to a tightly defined set of almost identical hydrophobic peptides from the highly conserved class l leader sequences. These molecular adaptations make HLA-E a rigorous checkpoint at the cell surface reporting on the integrity of the antigen processing pathway to CD94/NKG2 receptor-bearing natural killer cells.
- Nuffield Department of Clinical Medicine, University of Oxford, John Radcliffe Hospital, United Kingdom.
Organizational Affiliation: