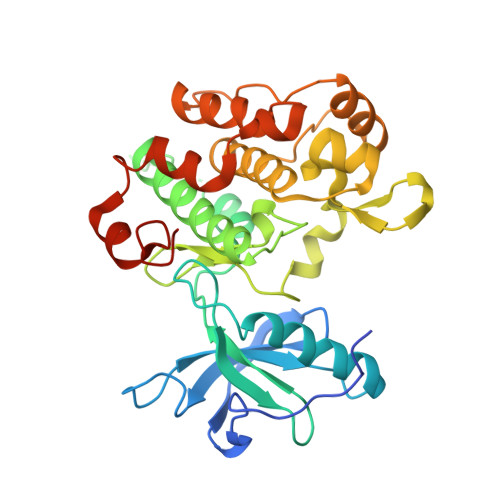
Crystal structure of the Apo, unactivated insulin-like growth factor-1 receptor kinase. Implication for inhibitor specificity.
Munshi, S., Kornienko, M., Hall, D.L., Reid, J.C., Waxman, L., Stirdivant, S.M., Darke, P.L., Kuo, L.C.(2002) J Biological Chem 277: 38797-38802
- PubMed: 12138114
- DOI: https://doi.org/10.1074/jbc.M205580200
- Primary Citation of Related Structures:
1M7N - PubMed Abstract:
The x-ray structure of the unactivated kinase domain of insulin-like growth factor-1 receptor (IGFRK-0P) is reported here at 2.7 A resolution. IGFRK-0P is composed of two lobes connected by a hinge region. The N-terminal lobe of the kinase is a twisted beta-sheet flanked by a single helix, and the C-terminal lobe comprises eight alpha-helices and four short beta-strands. The ATP binding pocket and the catalytic center reside at the interface of the two lobes. Despite the overall similarity to other receptor tyrosine kinases, three notable conformational modifications are observed: 1) this kinase adopts a more closed structure, with its two lobes rotated further toward each other; 2) the conformation of the proximal end of the activation loop (residues 1121-1129) is different; 3) the orientation of the nucleotide-binding loop is altered. Collectively, these alterations lead to a different ATP-binding pocket that might impact on inhibitor designs for IGFRK-0P. Two molecules of IGFRK-0P are seen in the asymmetric unit; they are associated as a dimer with their ATP binding clefts facing each other. The ordered N terminus of one monomer approaches the active site of the other, suggesting that the juxtamembrane region of one molecule could come into close proximity to the active site of the other.
- Department of Structural Biology, Merck Research Laboratories, West Point, Pennsylvania 19486, USA.
Organizational Affiliation: