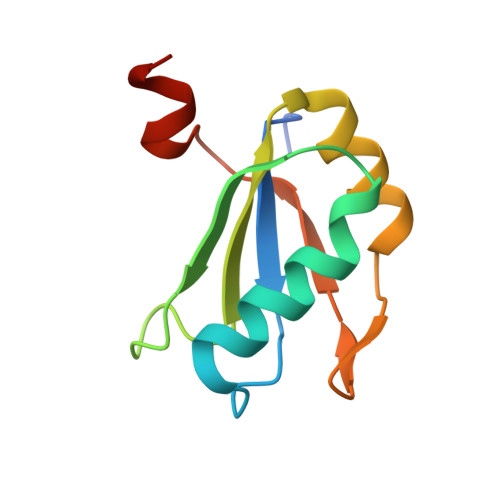
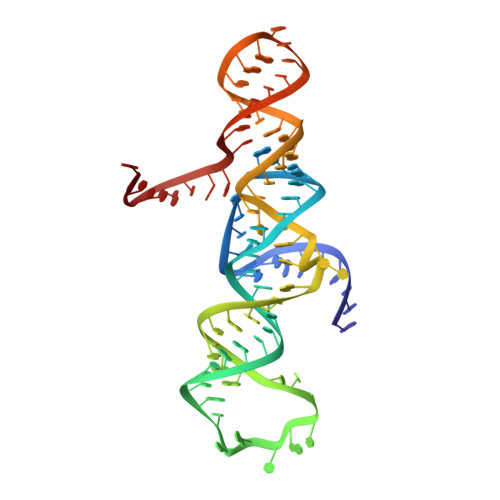
Transition state stabilization by a catalytic RNA
Rupert, P.B., Massey, A.P., Sigurdsson, S.T., Ferre-D'Amare, A.R.(2002) Science 298: 1421-1424
- PubMed: 12376595
- DOI: https://doi.org/10.1126/science.1076093
- Primary Citation of Related Structures:
1M5O, 1M5P, 1M5V - PubMed Abstract:
The hairpin ribozyme catalyzes sequence-specific cleavage of RNA through transesterification of the scissile phosphate. Vanadate has previously been used as a transition state mimic of protein enzymes that catalyze the same reaction. Comparison of the 2.2 angstrom resolution structure of a vanadate-hairpin ribozyme complex with structures of precursor and product complexes reveals a rigid active site that makes more hydrogen bonds to the transition state than to the precursor or product. Because of the paucity of RNA functional groups capable of general acid-base or electrostatic catalysis, transition state stabilization is likely to be an important catalytic strategy for ribozymes.
- Division of Basic Sciences, Fred Hutchinson Cancer Research Center, 1100 Fairview Avenue North, Seattle, WA 98109-1024, USA.
Organizational Affiliation: