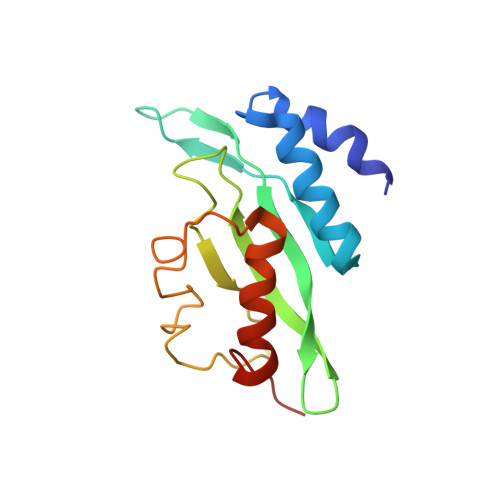
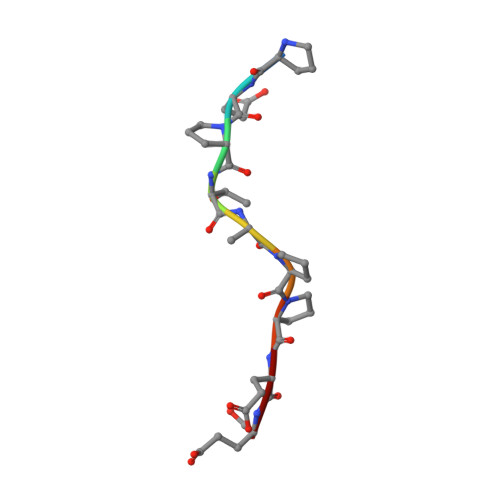
Structure of the Tsg101 UEV domain in complex with the PTAP motif of the HIV-1 p6 protein
Pornillos, O., Alam, S.L., Davis, D.R., Sundquist, W.I.(2002) Nat Struct Biol 9: 812-817
- PubMed: 12379843
- DOI: https://doi.org/10.1038/nsb856
- Primary Citation of Related Structures:
1M4P, 1M4Q - PubMed Abstract:
The structural proteins of HIV and Ebola display PTAP peptide motifs (termed 'late domains') that recruit the human protein Tsg101 to facilitate virus budding. Here we present the solution structure of the UEV (ubiquitin E2 variant) binding domain of Tsg101 in complex with a PTAP peptide that spans the late domain of HIV-1 p6(Gag). The UEV domain of Tsg101 resembles E2 ubiquitin-conjugating enzymes, and the PTAP peptide binds in a bifurcated groove above the vestigial enzyme active site. Each PTAP residue makes important contacts, and the Ala 9-Pro 10 dipeptide binds in a deep pocket of the UEV domain that resembles the X-Pro binding pockets of SH3 and WW domains. The structure reveals the molecular basis of HIV PTAP late domain function and represents an attractive starting point for the design of novel inhibitors of virus budding.
- Department of Biochemistry, University of Utah, Salt Lake City, Utah 84132, USA.
Organizational Affiliation: