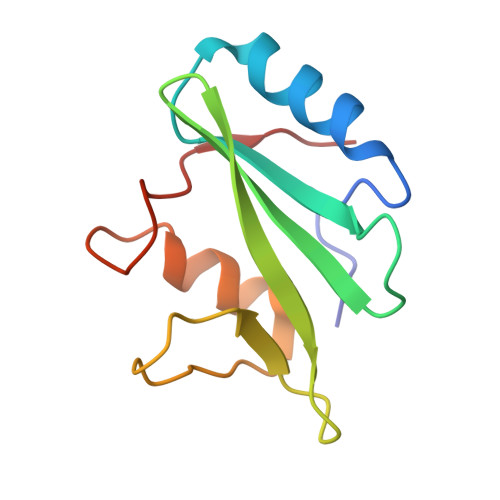
SAP couples Fyn to SLAM immune receptors.
Chan, B., Lanyi, A., Song, H.K., Griesbach, J., Simarro-Grande, M., Poy, F., Howie, D., Sumegi, J., Terhorst, C., Eck, M.J.(2003) Nat Cell Biol 5: 155-160
- PubMed: 12545174
- DOI: https://doi.org/10.1038/ncb920
- Primary Citation of Related Structures:
1M27 - PubMed Abstract:
SAP (SLAM-associated protein) is a small lymphocyte-specific signalling molecule that is defective or absent in patients with X-linked lymphoproliferative syndrome (XLP). Consistent with its single src homology 2 (SH2) domain architecture and unusually high affinity for SLAM (also called CD150), SAP has been suggested to function by blocking binding of SHP-2 or other SH2-containing signalling proteins to SLAM receptors. Additionally, SAP has recently been shown to be required for recruitment and activation of the Src-family kinase FynT after SLAM ligation. This signalling 'adaptor' function has been difficult to conceptualize, because unlike typical SH2-adaptor proteins, SAP contains only a single SH2 domain and lacks other recognized protein interaction domains or motifs. Here, we show that the SAP SH2 domain binds to the SH3 domain of FynT and directly couples FynT to SLAM. The crystal structure of a ternary SLAM-SAP-Fyn-SH3 complex reveals that SAP binds the FynT SH3 domain through a surface-surface interaction that does not involve canonical SH3 or SH2 binding interactions. The observed mode of binding to the Fyn-SH3 domain is expected to preclude the auto-inhibited conformation of Fyn, thereby promoting activation of the kinase after recruitment. These findings broaden our understanding of the functional repertoire of SH3 and SH2 domains.
- Department of Cancer Biology, Dana-Farber Cancer Institute, 44 Binney Street, Boston, MA 02115, USA.
Organizational Affiliation: