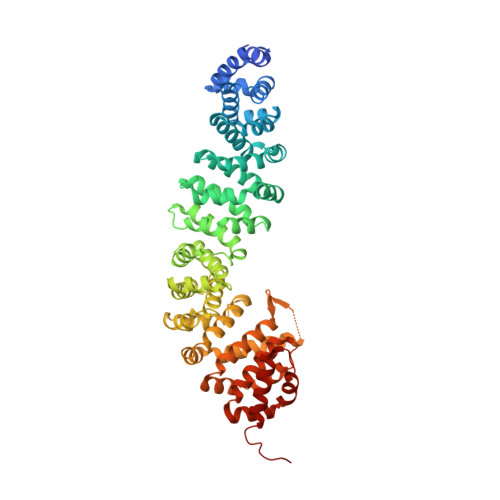
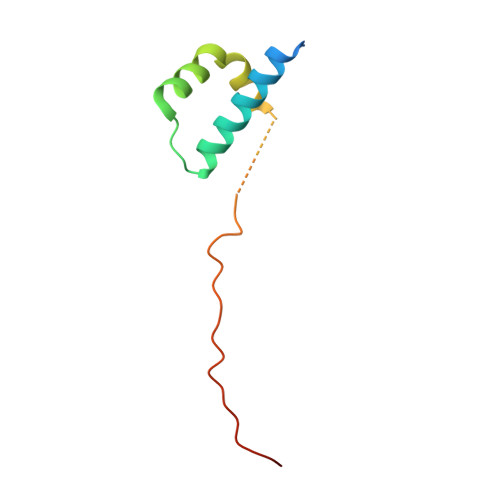
ICAT inhibits Beta-catenin binding to Tcf/Lef-family transcription factors and the general coactivator p300 using independent structural modules.
Daniels, D.L., Weis, W.I.(2002) Mol Cell 10: 573-584
- PubMed: 12408825
- DOI: https://doi.org/10.1016/s1097-2765(02)00631-7
- Primary Citation of Related Structures:
1M1E - PubMed Abstract:
In the canonical Wnt signaling pathway, beta-catenin activates target genes through its interactions with Tcf/Lef-family transcription factors and additional transcriptional coactivators. The crystal structure of ICAT, an inhibitor of beta-catenin-mediated transcription, bound to the armadillo repeat domain of beta-catenin, has been determined. ICAT contains an N-terminal helilical domain that binds to repeats 11 and 12 of beta-catenin, and an extended C-terminal region that binds to repeats 5-10 in a manner similar to that of Tcfs and other beta-catenin ligands. Full-length ICAT dissociates complexes of beta-catenin, Lef-1, and the transcriptional coactivator p300, whereas the helical domain alone selectively blocks binding to p300. The C-terminal armadillo repeats of beta-catenin may be an attractive target for compounds designed to disrupt aberrant beta-catenin-mediated transcription associated with various cancers.
- Department of Structural Biology, Stanford University School of Medicine, Stanford, CA 94305, USA.
Organizational Affiliation: