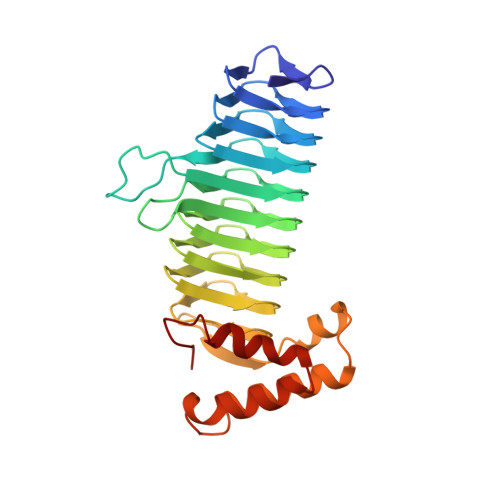
A left-handed parallel beta helix in the structure of UDP-N-acetylglucosamine acyltransferase.
Raetz, C.R., Roderick, S.L.(1995) Science 270: 997-1000
- PubMed: 7481807
- DOI: https://doi.org/10.1126/science.270.5238.997
- Primary Citation of Related Structures:
1LXA - PubMed Abstract:
UDP-N-acetylglucosamine 3-O-acyltransferase (LpxA) catalyzes the transfer of (R)-3-hydroxymyristic acid from its acyl carrier protein thioester to UDP-N-acetylglucosamine. LpxA is the first enzyme in the lipid A biosynthetic pathway and is a target for the design of antibiotics. The x-ray crystal structure of LpxA has been determined to 2.6 angstrom resolution and reveals a domain motif composed of parallel beta strands, termed a left-handed parallel beta helix (L beta H). This unusual fold displays repeated violations of the protein folding constraint requiring right-handed crossover connections between strands of parallel beta sheets and may be present in other enzymes that share amino acid sequence homology to the repeated hexapeptide motif of LpxA.
- Department of Biochemistry, Duke University Medical Center, Durham, NC 22710, USA.
Organizational Affiliation: