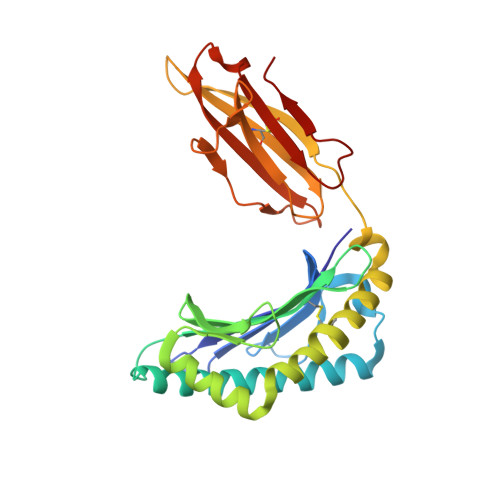
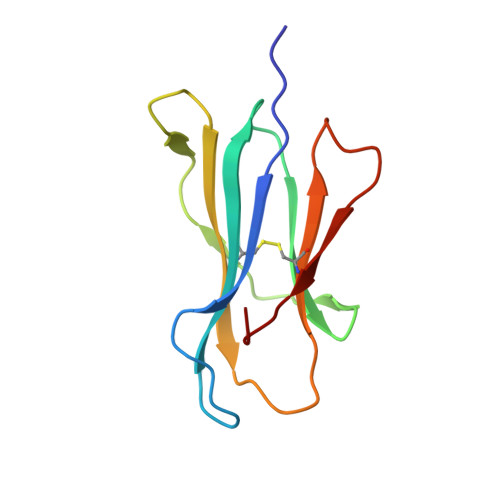
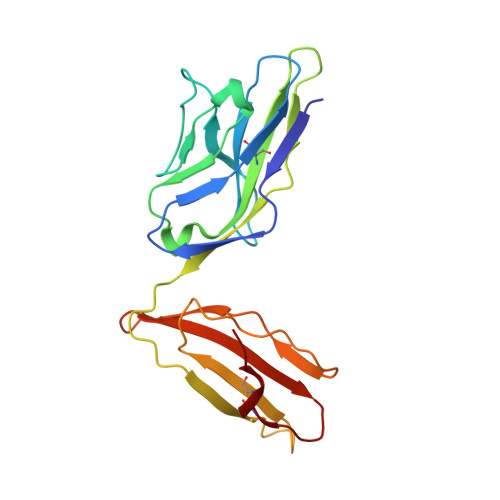
A correlation between TCR Valpha docking on MHC and CD8 dependence: implications for T cell selection.
Buslepp, J., Wang, H., Biddison, W.E., Appella, E., Collins, E.J.(2003) Immunity 19: 595-606
- PubMed: 14563323
- DOI: https://doi.org/10.1016/s1074-7613(03)00269-3
- Primary Citation of Related Structures:
1LP9 - PubMed Abstract:
T cell receptors (TCR) adopt a similar orientation when binding with major histocompatibility complex (MHC) molecules, yet the biological mechanism that generates this similar TCR orientation remains obscure. We show here the cocrystallographic structure of a mouse TCR bound to a human MHC molecule not seen by the TCR during thymic development. The orientation of this xenoreactive murine TCR atop human MHC deviates from the typical orientation more than any previously determined TCR/MHC structure. This unique orientation is solely due to the placement of the TCR Valpha domain on the MHC. In light of new information provided by this structure, we have reanalyzed the existing TCR/MHC cocrystal structures and discovered unique features of TCR Valpha domain position on class I MHC that correlate with CD8 dependence. Finally, we propose that the orientation seen in TCR recognition of MHC is a consequence of selection during T cell development.
- Department of Biochemistry and Biophysics, University of North Carolina, Chapel Hill, NC 27599, USA.
Organizational Affiliation: