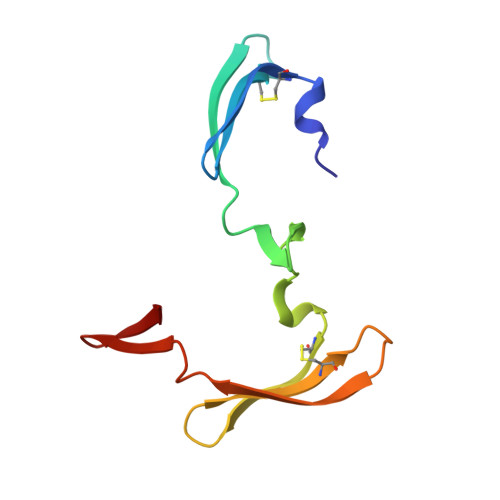
Domain-swapped structure of a mutant of cyanovirin-N.
Botos, I., Mori, T., Cartner, L.K., Boyd, M.R., Wlodawer, A.(2002) Biochem Biophys Res Commun 294: 184-190
- PubMed: 12054761
- DOI: https://doi.org/10.1016/S0006-291X(02)00455-2
- Primary Citation of Related Structures:
1LOM - PubMed Abstract:
Cyanovirin-N (CV-N) is a potent 11 kDa HIV-inactivating protein that binds with high affinity to the HIV surface envelope protein gp120. A double mutant P51S/S52P of CV-N was engineered by swapping two critical hinge-region residues Pro51 and Ser52. This mutant has biochemical and biophysical characteristics equivalent to the wild-type CV-N and its structure resembles that of wild-type CV-N. However, the mutant shows a different orientation in the hinge region that connects two domains of the protein. The observation that this double mutant crystallizes under a wide variety of conditions challenges some of the current hypotheses on domain swapping and on the role of hinge-region proline residues in domain orientation. The current structure contributes to the understanding of domain swapping in cyanovirins, permitting rational design of domain-swapped CV-N mutants.
- Macromolecular Crystallography Laboratory, National Cancer Institute at Frederick, Frederick, MD 21702-1201, USA.
Organizational Affiliation: