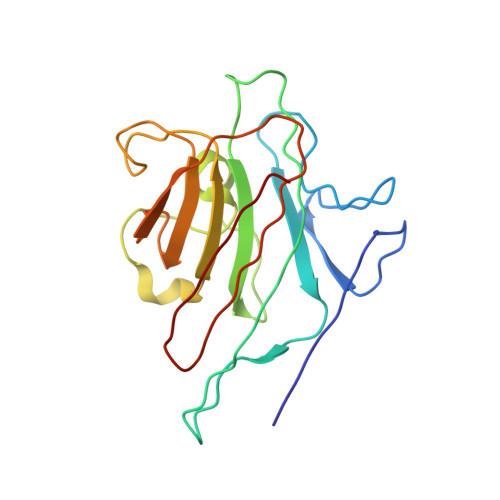
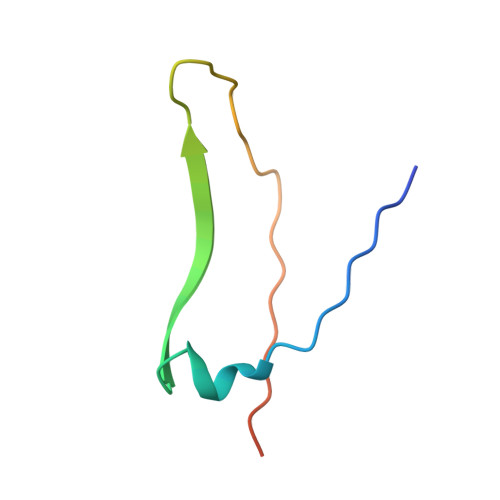
X-ray structure of a (alpha-Man(1-3)beta-Man(1-4)GlcNAc)-lectin complex at 2.1-A resolution. The role of water in sugar-lectin interaction.
Bourne, Y., Rouge, P., Cambillau, C.(1990) J Biological Chem 265: 18161-18165
- PubMed: 2211692
- Primary Citation of Related Structures:
1LOG - PubMed Abstract:
We describe herein the high resolution refined x-ray structure of a trisaccharide, which is a part of the N-acetyllactosamine type glycan found in the majority of the N-glycosyl-proteins, complexed to the isolectin I. According to the potentials used by Imberty et al. (Imburty, A., Gerber, S., Tran, V., and Pérez, S. (1990) Glycoconjugate J. 7, 27-54) the trisaccharide is in a low-energy state. Only one mannose moiety establishes direct hydrogen bonds with the lectin, as it is the case for monosaccharide-lectin complexes. The comparison of our trisaccharide with the one determined in solution by Warin et al. (Warin, V., Baert, F., Fouret, R., Strecker, G., Fournet, B., and Montreuil, J. (1979) Carbohydr. Res. 76, 11-22) shows that both adopt roughly the same conformation. The differences in these two sugar structures allow us to assign the role of water molecules present in the vicinity of our trisaccharide for the stabilization of this sugar-lectin complex.
- Laboratoire de Cristallographie et de Cristallisation des Macromolécules Biologiques, Faculté de Médecine, Marseille, France.
Organizational Affiliation: