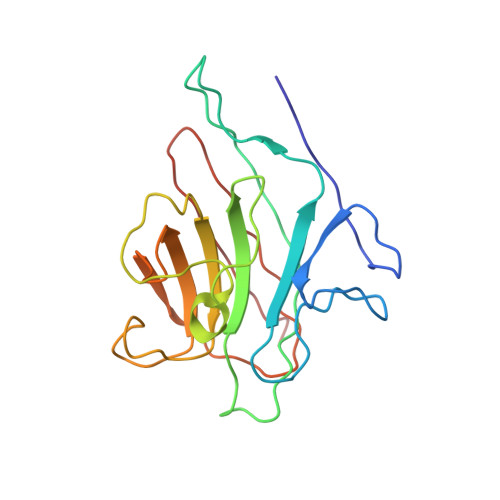
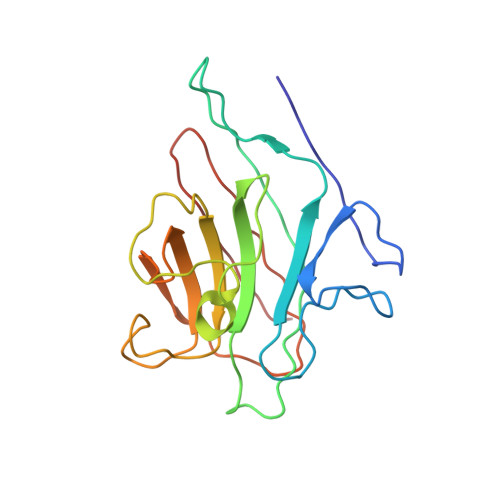
X-ray structure of a biantennary octasaccharide-lectin complex refined at 2.3-A resolution.
Bourne, Y., Rouge, P., Cambillau, C.(1992) J Biological Chem 267: 197-203
- PubMed: 1730588
- Primary Citation of Related Structures:
1LOF - PubMed Abstract:
The structure and flexibility of saccharides have a profound and specific influence in several biological processes such as protein protection and the maintenance of conformational integrity, and in recognition events involving viruses, enzymes, and lectins. To establish the structural bases of these phenomena, we describe herein the extensively refined 2.3-A resolution x-ray structure of a biantennary octasaccharide of the N-acetyllactosamine type, complexed to isolectin I from Lathyrus ochrus. The two octasaccharides are located in clefts at each end of the long axis of the lectin. The conformations of both the lectin and the saccharide are slightly modified upon binding. The complex is stabilized by numerous hydrogen bonds, many of them involving water molecules. It is also stabilized by van der Waals interactions, including some with aromatic residues. A more general model of a possible lectin-glycoprotein interaction is also proposed.
- Laboratoire de Cristallographie et de Cristallisation des Macromolécules Biologiques, Faculté de Médecine Secteur-Nord, Marseille, France.
Organizational Affiliation: