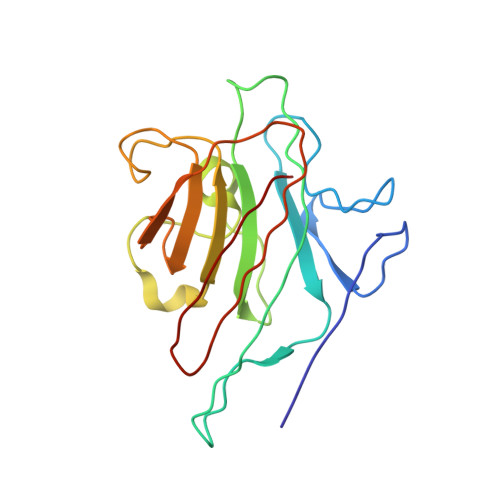
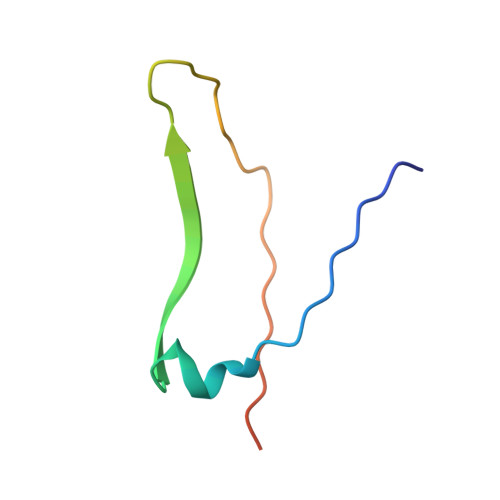
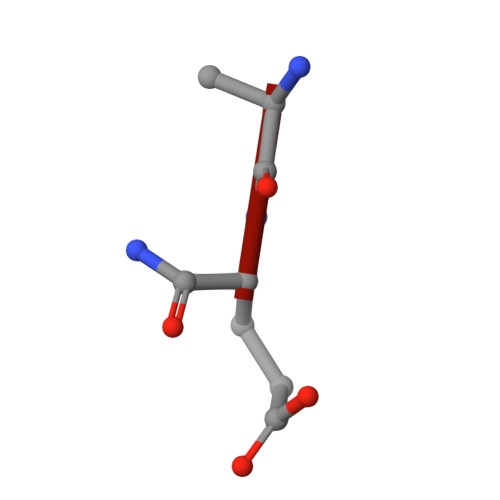
Interaction of a legume lectin with two components of the bacterial cell wall. A crystallographic study.
Bourne, Y., Ayouba, A., Rouge, P., Cambillau, C.(1994) J Biological Chem 269: 9429-9435
- PubMed: 8144527
- Primary Citation of Related Structures:
1LOC, 1LOD - PubMed Abstract:
We describe herein the refined high resolution x-ray structures of two components of the bacterial cell wall, muramic acid and muramyl dipeptide complexed to isolectin I from Lathyrus ochrus seeds. In both complexes, only the ring hydroxyl oxygen atoms of the bound sugar establish direct hydrogen bonds with isolectin I, as in the case of all the previously determined monosaccharide-lectin complexes. In addition, the lactyl methyl of both components strongly interacts via hydrophobic contacts with the side chains of residues Tyr100 and Trp128 of isolectin I, which could explain the higher affinity of isolectin I for muramic acid as compared with glucose. These 2 residues, however, are not involved in the stabilization of the oligosaccharide-isolectin I complexes. The dipeptide (D-Ala-D-iGln) of the second component is in stacking interaction with the N-acetyl group of glucose and with loop Gly97-Gly98 of isolectin I. In addition to these van der Waals' contacts, the dipeptide interacts with the lectin via well ordered water molecules also. Superposition of the structures of the muramyl dipeptide complex and of the muramic acid complex shows that the glucose ring in the dipeptide compound is tilted by about 15 degrees in comparison with that of muramic acid. The fact that the lactyl group has the same confrontation in both components reveals that the lectin is stereospecific and recognizes only diastereoisomer S of this group, which better fits the saccharide-binding site.
- Laboratorie de Cristallographie et de Cristallisation des Macromolécules Biologiques, Centre National de la Recherche Scientifique Unité de Recherche associée 1296, Faculté de Médecine Secteur-Nord, Marseille, France.
Organizational Affiliation: