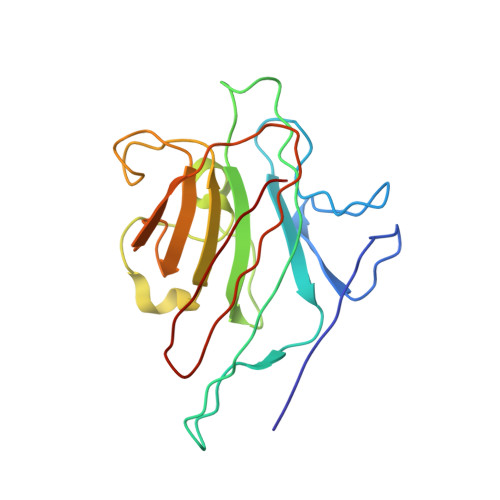
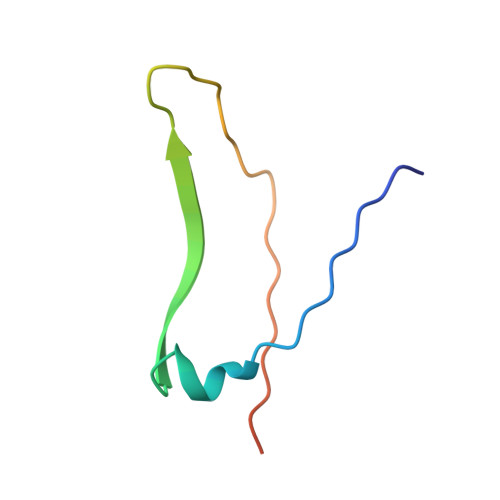
Three-dimensional structures of complexes of Lathyrus ochrus isolectin I with glucose and mannose: fine specificity of the monosaccharide-binding site.
Bourne, Y., Roussel, A., Frey, M., Rouge, P., Fontecilla-Camps, J.C., Cambillau, C.(1990) Proteins 8: 365-376
- PubMed: 2091026
- DOI: https://doi.org/10.1002/prot.340080410
- Primary Citation of Related Structures:
1LOA, 1LOB - PubMed Abstract:
The structure of the methyl-alpha-D-mannopyranoside-LOL I complex has been solved by the molecular replacement method using the refined saccharide-free LOL I coordinates as starting model. The methyl-alpha-D-mannopyranoside-LOL I complex was refined by simulated annealing using the program X-PLOR. The final R-factor value is 0.182 [Fo greater than 1 sigma(Fo)]. The isostructural methyl-alpha-D-glucopyranoside-LOL I complex was refined by X-Ray coupled energy minimization using the methyl-alpha-D-mannopyranoside-LOL I structure as a starting model to an R factor of 0.179 (all data). In both crystal forms, each dimer binds two molecules of sugar in pockets found near the calcium ions. The two saccharide moieties, which are in the C1 chair conformation, establish the same hydrogen bond pattern with the lectin. However, the van der Waals contacts are different between the O2, C2, C6, and O6 atoms of the two molecules and the backbone atoms of residues 208-211. Mannose, due to its axial C2 conformation, encloses the backbone atoms of the protein in a clamplike way. Van der Waals energy calculations suggest that this better complementarity of the mannoside molecule with the lectin could explain its higher affinity for isolectin I.
- Laboratoire de Cristallographie et de Cristallisation des Macromolécules Biologiques, Faculté de Médecine, Marseille, France.
Organizational Affiliation: