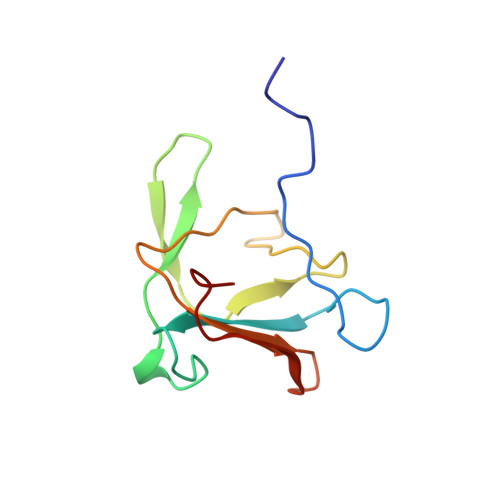
Phage P22 tailspike protein: crystal structure of the head-binding domain at 2.3 A, fully refined structure of the endorhamnosidase at 1.56 A resolution, and the molecular basis of O-antigen recognition and cleavage.
Steinbacher, S., Miller, S., Baxa, U., Budisa, N., Weintraub, A., Seckler, R., Huber, R.(1997) J Mol Biology 267: 865-880
- PubMed: 9135118
- DOI: https://doi.org/10.1006/jmbi.1997.0922
- Primary Citation of Related Structures:
1LKT - PubMed Abstract:
The tailspike protein of Salmonella phage P22 is a viral adhesion protein with both receptor binding and destroying activities. It recognises the O-antigenic repeating units of cell surface lipopolysaccharide of serogroup A, B and D1 as receptor, but also inactivates its receptor by endoglycosidase (endorhamnosidase) activity. In the final step of bacteriophage P22 assembly six homotrimeric tailspike molecules are non-covalently attached to the DNA injection apparatus, mediated by their N-terminal, head-binding domains. We report the crystal structure of the head-binding domain of P22 tailspike protein at 2.3 A resolution, solved with a recombinant telluromethionine derivative and non-crystallographic symmetry averaging. The trimeric dome-like structure is formed by two perpendicular beta-sheets of five and three strands, respectively in each subunit and caps a three-helix bundle observed in the structure of the C-terminal receptor binding and cleaving fragment, reported here after full refinement at 1.56 A resolution. In the central part of the receptor binding fragment, three parallel beta-helices of 13 complete turns are associated side-by-side, while the three polypeptide strands merge into a single domain towards their C termini, with close interdigitation at the junction to the beta-helix part. Complex structures with receptor fragments from S. typhimurium, S. enteritidis and S. typhi253Ty determined at 1.8 A resolution are described in detail. Insertions into the beta-helix form the O-antigen binding groove, which also harbours the active site residues Asp392, Asp395 and Glu359. In the intact structure of the tailspike protein, head-binding and receptor-binding parts are probably linked by a flexible hinge whose function may be either to deal with shearing forces on the exposed, 150 A long tailspikes or to allow them to bend during the infection process.
- Max-Planck-Institut für Biochemie, Abteilung für Strukturforschung, Martinsried, Germany.
Organizational Affiliation: