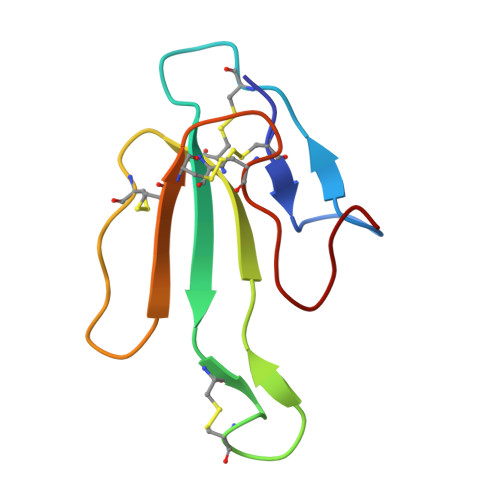
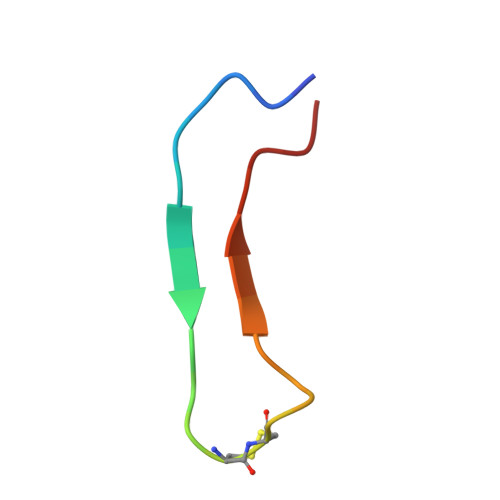
The mechanism for acetylcholine receptor inhibition by alpha-neurotoxins and species-specific resistance to alpha-bungarotoxin revealed by NMR.
Samson, A., Scherf, T., Eisenstein, M., Chill, J., Anglister, J.(2002) Neuron 35: 319-332
- PubMed: 12160749
- DOI: https://doi.org/10.1016/s0896-6273(02)00773-0
- Primary Citation of Related Structures:
1L4W, 1LJZ - PubMed Abstract:
The structure of a peptide corresponding to residues 182-202 of the acetylcholine receptor alpha1 subunit in complex with alpha-bungarotoxin was solved using NMR spectroscopy. The peptide contains the complete sequence of the major determinant of AChR involved in alpha-bungarotoxin binding. One face of the long beta hairpin formed by the AChR peptide consists of exposed nonconserved residues, which interact extensively with the toxin. Mutations of these receptor residues confer resistance to the toxin. Conserved AChR residues form the opposite face of the beta hairpin, which creates the inner and partially hidden pocket for acetylcholine. An NMR-derived model for the receptor complex with two alpha-bungarotoxin molecules shows that this pocket is occupied by the conserved alpha-neurotoxin residue R36, which forms cation-pi interactions with both alphaW149 and gammaW55/deltaW57 of the receptor and mimics acetylcholine.
- Department of Structural Biology, The Weizmann Institute of Science, Rehovot, Israel.
Organizational Affiliation: