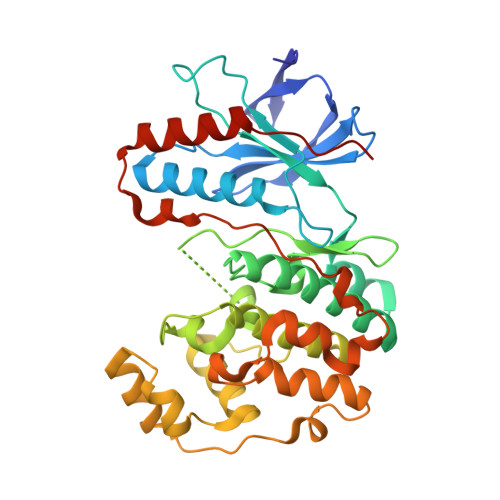
Crystal structures of MAP kinase p38 complexed to the docking sites on its nuclear substrate MEF2A and activator MKK3b.
Chang, C.I., Xu, B.E., Akella, R., Cobb, M.H., Goldsmith, E.J.(2002) Mol Cell 9: 1241-1249
- PubMed: 12086621
- DOI: https://doi.org/10.1016/s1097-2765(02)00525-7
- Primary Citation of Related Structures:
1LEW, 1LEZ - PubMed Abstract:
The structures of the MAP kinase p38 in complex with docking site peptides containing a phi(A)-X-phi(B) motif, derived from substrate MEF2A and activating enzyme MKK3b, have been solved. The peptides bind to the same site in the C-terminal domain of the kinase, which is both outside the active site and distinct from the "CD" domain previously implicated in docking site interactions. Mutational analysis on the interaction of p38 with the docking sites supports the crystallographic models and has uncovered two novel residues on the docking groove that are critical for binding. The two peptides induce similar large conformational changes local to the peptide binding groove. The peptides also induce unexpected and different conformational changes in the active site, as well as structural disorder in the phosphorylation lip.
- Department of Biochemistry, The University of Texas Southwestern Medical Center at Dallas, 5323 Harry Hines Boulevard, Dallas, TX 75390, USA.
Organizational Affiliation: