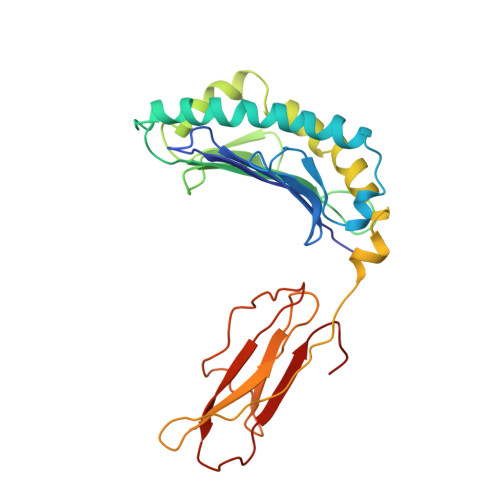
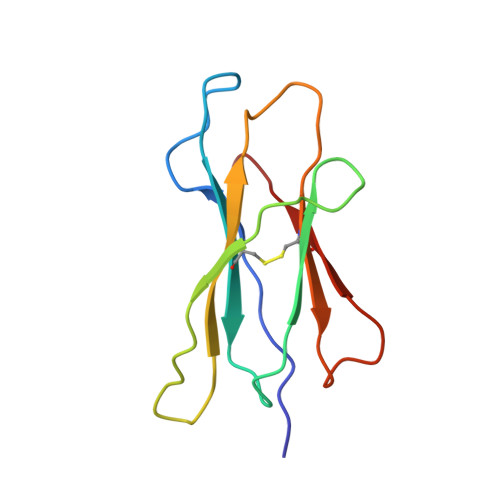
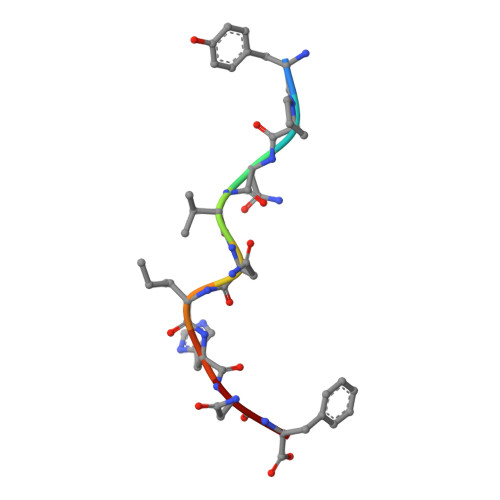
The three-dimensional structure of an H-2Ld-peptide complex explains the unique interaction of Ld with beta-2 microglobulin and peptide.
Balendiran, G.K., Solheim, J.C., Young, A.C., Hansen, T.H., Nathenson, S.G., Sacchettini, J.C.(1997) Proc Natl Acad Sci U S A 94: 6880-6885
- PubMed: 9192660
- DOI: https://doi.org/10.1073/pnas.94.13.6880
- Primary Citation of Related Structures:
1LD9 - PubMed Abstract:
Solution at 2.5-A resolution of the three-dimensional structure of H-2Ld with a single nine-residue peptide provides a structural basis for understanding its unique interaction with beta-2 microglobulin (beta2m) and peptide. Consistent with the biological data that show an unusually weak association of Ld with beta2m, a novel orientation of the alpha1/alpha2 domains of Ld relative to beta2m results in a dearth of productive contacts compared with other class I proteins. Characteristics of the Ld antigen-binding cleft determine the unique motif of peptides that it binds. Ld has no central anchor residue due to the presence of several bulky side chains in its mid-cleft region. Also, its cleft is significantly more hydrophobic than that of the other class I molecules for which structures are known, resulting in many fewer H-bonds between peptide and cleft residues. The choice of Pro as a consensus anchor at peptide position 2 appears to be related to the hydrophobicity of the B pocket, and to the unique occurrence of Ile (which mirrors Pro in its inability to form H-bonds) at position 63 on the edge of this pocket. Thus, the paucity of stabilizing H-bonds combined with poor complementarity between peptide postion 2 Pro and the B pocket contribute to the weak association between Ld and its peptide antigen. The unique structural interactions of Ld with beta2m and peptide could make Ld more suited than other classical class I molecules to play a role in alternative pathways of antigen presentation.
- Department of Biochemistry and Biophysics, Texas A&M University, College Station, TX 77843, USA.
Organizational Affiliation: