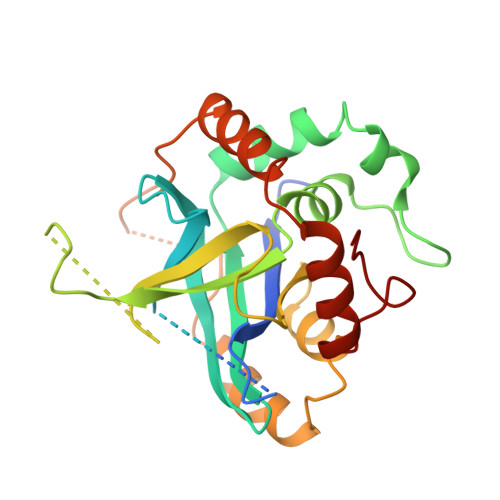
Unique fold and active site in cytomegalovirus protease.
Qiu, X., Culp, J.S., DiLella, A.G., Hellmig, B., Hoog, S.S., Janson, C.A., Smith, W.W., Abdel-Meguid, S.S.(1996) Nature 383: 275-279
- PubMed: 8805707
- DOI: https://doi.org/10.1038/383275a0
- Primary Citation of Related Structures:
1LAY - PubMed Abstract:
Human herpesviruses are responsible for a variety of diseases. They are divided into three subfamilies: alpha includes herpes simplex viruses (HSV-1 and HSV-2) and varicella-zoster virus (VZV); beta includes cytomegalovirus (CMV) and human herpesvirus-6 (HHV-6); and gamma includes Epstein-Barr virus (EBV). Each virus encodes a serine protease that is essential for its replication and is a potential target for therapeutic intervention. Human CMV is a ubiquitous opportunistic pathogen that can result in life-threatening infections in congenitally infected infants, immunocompromised individuals and immunosuppressed cancer or transplant patients. Here we report the crystal structure of human CMV protease at 2.5 angstroms resolution. The structure reveals a fold that has not been reported for any other serine protease, and an active site consisting of a novel catalytic triad in which the third member is a histidine instead of an aspartic acid, or possibly a catalytic tetrad consisting of a serine, two histidines and an aspartic acid. An unusual dimer interface that is important to the protease activity has also been identified.
- Department of Macromolecular Sciences, SmithKline Beecham Pharmaceuticals, King of Prussia, Pennsylvania 19406, USA.
Organizational Affiliation: