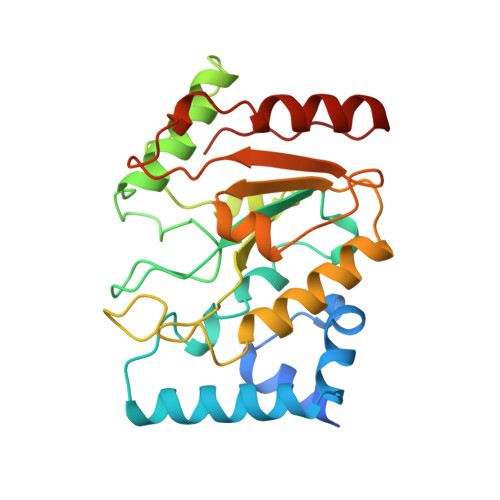
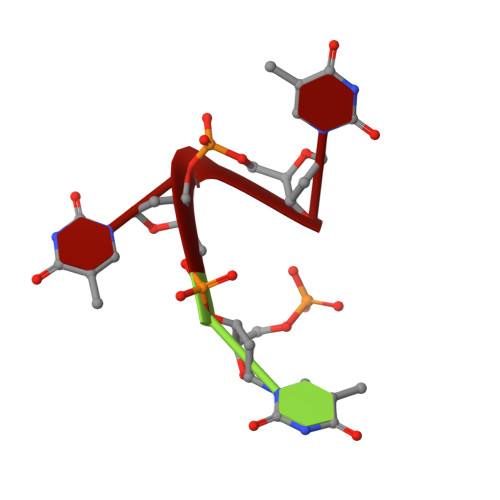
The structural basis of specific base-excision repair by uracil-DNA glycosylase.
Savva, R., McAuley-Hecht, K., Brown, T., Pearl, L.(1995) Nature 373: 487-493
- PubMed: 7845459
- DOI: https://doi.org/10.1038/373487a0
- Primary Citation of Related Structures:
1LAU, 1UDG, 1UDH - PubMed Abstract:
The 1.75-A crystal structure of the uracil-DNA glycosylase from herpes simplex virus type-1 reveals a new fold, distantly related to dinucleotide-binding proteins. Complexes with a trideoxynucleotide, and with uracil, define the DNA-binding site and allow a detailed understanding of the exquisitely specific recognition of uracil in DNA. The overall structure suggests binding models for elongated single- and double-stranded DNA substrates. Conserved residues close to the uracil-binding site suggest a catalytic mechanism for hydrolytic base excision.
- Department of Biochemistry and Molecular Biology, University College London, UK.
Organizational Affiliation: