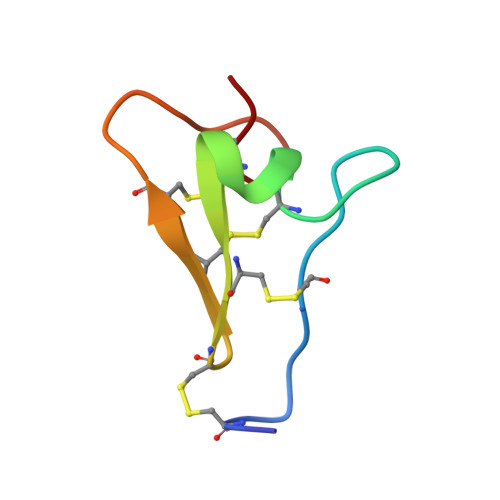
Cysteine-rich module structure reveals a fulcrum for integrin rearrangement upon activation.
Beglova, N., Blacklow, S.C., Takagi, J., Springer, T.A.(2002) Nat Struct Biol 9: 282-287
- PubMed: 11896403
- DOI: https://doi.org/10.1038/nsb779
- Primary Citation of Related Structures:
1L3Y - PubMed Abstract:
Cysteine-rich repeats in the integrin beta subunit stalk region relay activation signals to the ligand-binding headpiece. The NMR solution structure and disulfide bond connectivity of Cys-rich module-3 of the integrin beta2 subunit reveal a nosecone-shaped variant of the EGF fold, termed an integrin-EGF (I-EGF) domain. Interdomain contacts between I-EGF domains 2 and 3 observed by NMR support a model in which the modules are related by an approximate two-fold screw axis in an extended arrangement. Our findings complement a 3.1 A crystal structure of the extracellular portion of integrin alphaVbeta3, which lacks an atomic model for I-EGF2 and a portion of I-EGF3. The disulfide connectivity of I-EGF3 chemically assigned here differs from the pairings suggested in the alphaVbeta3 structure. Epitopes that become exposed upon integrin activation and residues that restrain activation are defined in beta2 I-EGF domains 2 and 3. Superposition on the alphaVbeta3 structure reveals that they are buried. This observation suggests that the highly bent alphaVbeta3 structure represents the inactive conformation and that release of contacts with I-EGF modules 2 and 3 triggers a switchblade-like opening motion extending the integrin into its active conformation.
- Department of Pathology, Harvard Medical School, 75 Francis St., Boston, Masschusetts 02115, USA.
Organizational Affiliation: