Optimization of the beta-aminoester class of factor Xa inhibitors. Part 2: Identification of FXV673 as a potent and selective inhibitor with excellent In vivo anticoagulant activity.
Guertin, K.R., Gardner, C.J., Klein, S.I., Zulli, A.L., Czekaj, M., Gong, Y., Spada, A.P., Cheney, D.L., Maignan, S., Guilloteau, J.P., Brown, K.D., Colussi, D.J., Chu, V., Heran, C.L., Morgan, S.R., Bentley, R.G., Dunwiddie, C.T., Leadley, R.J., Pauls, H.W.(2002) Bioorg Med Chem Lett 12: 1671-1674
- PubMed: 12039587
- DOI: https://doi.org/10.1016/s0960-894x(02)00213-5
- Primary Citation of Related Structures:
1KSN - PubMed Abstract:
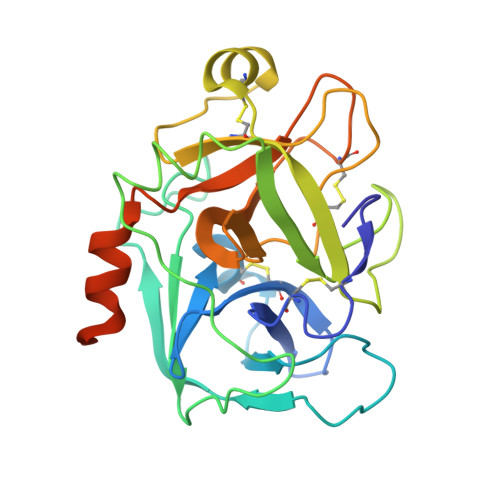
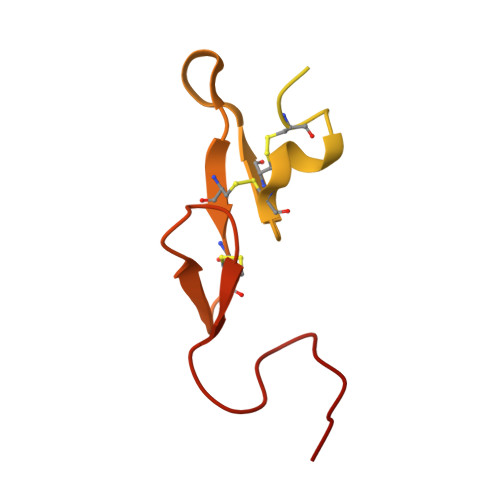
Further optimization of the beta-aminoester class of factor Xa (fXa) inhibitors is described culminating in the identification of 9c (FXV673), a potent and selective factor Xa inhibitor with excellent in vivo anticoagulant activity. An X-ray structure of FXV673 bound to human fXa is also presented. Based on its selectivity, potent in vivo activity and favorable pre-clinical safety profile, FXV673 was selected for further development and is currently undergoing clinical trials.
- Drug Innovation and Approval, Aventis Pharmaceuticals, Route 202-206, Bridgewater, NJ 08807, USA. kevin.guertin@roche.com
Organizational Affiliation: