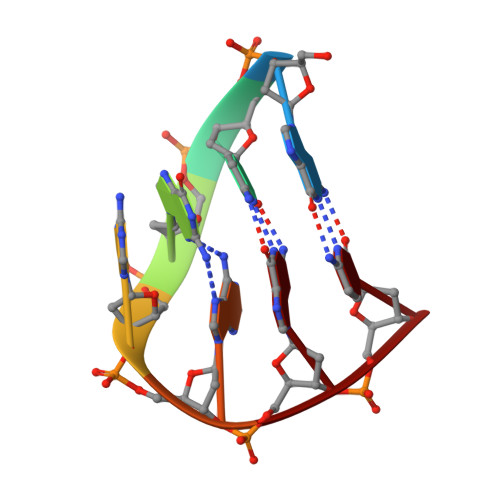
Refinement of d(GCGAAGC) Hairpin Structure Using One- and Two-Bond Residual Dipolar Couplings
Padrta, P., Stefl, R., Kralik, L., Zidek, L., Sklenar, V.(2002) J Biomol NMR 24: 1-14
- PubMed: 12449414
- DOI: https://doi.org/10.1023/a:1020632900961
- Primary Citation of Related Structures:
1KR8, 1PQT - PubMed Abstract:
The structure of the 13C,15N-labeled d(GCGAAGC) hairpin, as determined by NMR spectroscopy and refined using molecular dynamics with NOE-derived distances, torsion angles, and residual dipolar couplings (RDCs), is presented. Although the studied molecule is of small size, it is demonstrated that the incorporation of diminutive RDCs can significantly improve local structure determination of regions undefined by the conventional restraints. Very good correlation between the experimental and back-calculated small one- and two-bond 1H-13C, 1H-15N, 13C-13C and 13C-15N coupling constants has been attained. The final structures clearly show typical features of the miniloop architecture. The structure is discussed in context of the extraordinary stability of the d(GCGAAGC) hairpin, which originates from a complex interplay between the aromatic base stacking and hydrogen bonding interactions.
- National Centre for Biomolecular Research, Masaryk University, Brno, Czech Republic.
Organizational Affiliation: