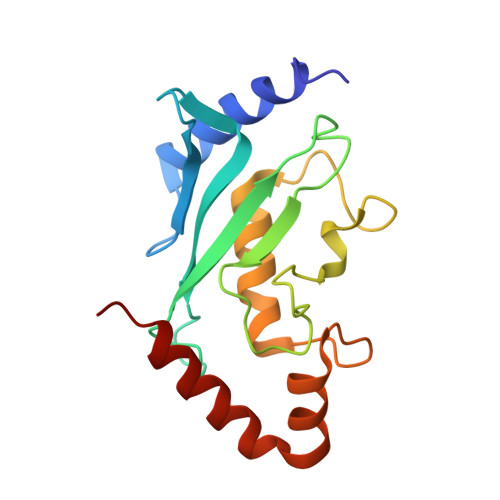
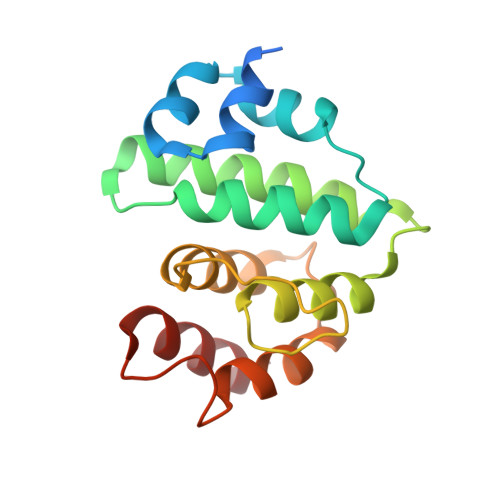
Structural basis for E2-mediated SUMO conjugation revealed by a complex between ubiquitin-conjugating enzyme Ubc9 and RanGAP1.
Bernier-Villamor, V., Sampson, D.A., Matunis, M.J., Lima, C.D.(2002) Cell 108: 345-356
- PubMed: 11853669
- DOI: https://doi.org/10.1016/s0092-8674(02)00630-x
- Primary Citation of Related Structures:
1KPS - PubMed Abstract:
E2 enzymes catalyze attachment of ubiquitin and ubiquitin-like proteins to lysine residues directly or through E3-mediated reactions. The small ubiquitin-like modifier SUMO regulates nuclear transport, stress response, and signal transduction in eukaryotes and is essential for cell-cycle progression in yeast. In contrast to most ubiquitin conjugation, the SUMO E2 enzyme Ubc9 is sufficient for substrate recognition and lysine modification of known SUMO targets. Crystallographic analysis of a complex between mammalian Ubc9 and a C-terminal domain of RanGAP1 at 2.5 A reveals structural determinants for recognition of consensus SUMO modification sequences found within SUMO-conjugated proteins. Structure-based mutagenesis and biochemical analysis of Ubc9 and RanGAP1 reveal distinct motifs required for substrate binding and SUMO modification of p53, IkappaBalpha, and RanGAP1.
- Biochemistry Department, Structural Biology Program, Weill Medical College of Cornell University, New York, NY 10021, USA.
Organizational Affiliation: