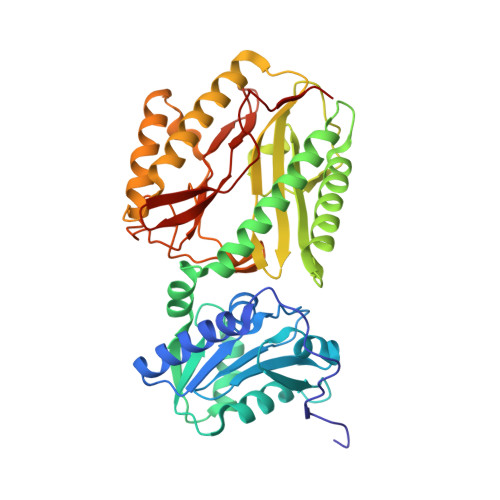
Structure of creatine amidinohydrolase from Actinobacillus.
Padmanabhan, B., Paehler, A., Horikoshi, M.(2002) Acta Crystallogr D Biol Crystallogr 58: 1322-1328
- PubMed: 12136144
- DOI: https://doi.org/10.1107/s0907444902010156
- Primary Citation of Related Structures:
1KP0 - PubMed Abstract:
The crystal structure of Actinobacillus creatine amidinohydrolase has been solved by molecular replacement. The amino-acid sequence has been derived from the crystal structure. Crystals belong to space group I222, with unit-cell parameters a = 111.26 (3), b = 113.62 (4), c = 191.65 (2) A, and contain two molecules in an asymmetric unit. The structure was refined to an R factor of 18.8% at 2.7 A resolution. The crystal structure contains a dimer of 402 residues and 118 water molecules. The protein structure is bilobal, consisting of a small N-terminal domain and a large C-terminal domain. The C-terminal domain has a pitta-bread fold, similar to that found in Pseudomonas putida creatinase, proline aminopeptidases and methionine aminopeptidase. Comparison with complex crystal structures of P. putida creatinase reveals that the enzyme activity of Actinobacillus creatinase might be similar to that of P. putida creatinase.
- Horikoshi Gene Selector Project, Exploratory Research for Advanced Technology (ERATO), Japan Science and Technology Corporation (JST), 5-9-6 Tokodai, Tsukuba, Ibaraki 300-2635, Japan.
Organizational Affiliation: