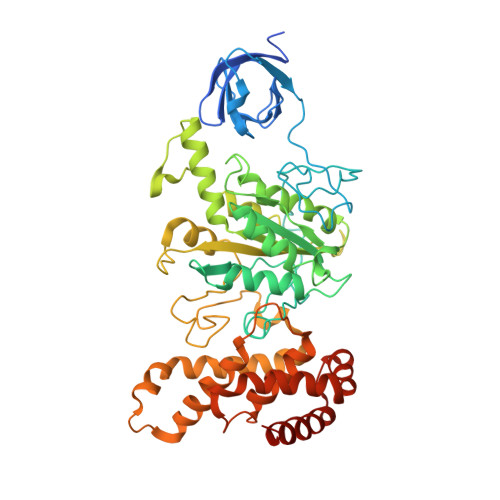
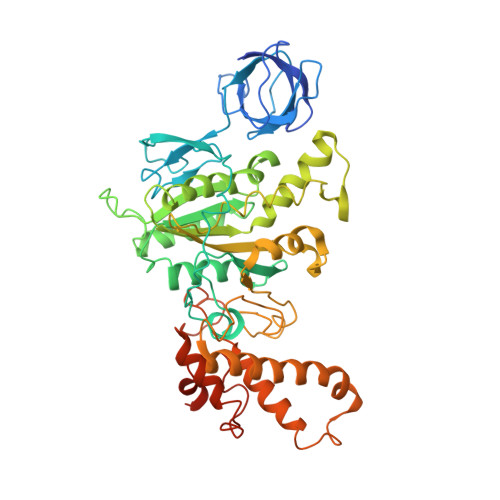
Structure of spinach chloroplast F1-ATPase complexed with the phytopathogenic inhibitor tentoxin.
Groth, G.(2002) Proc Natl Acad Sci U S A 99: 3464-3468
- PubMed: 11904410
- DOI: https://doi.org/10.1073/pnas.052546099
- Primary Citation of Related Structures:
1KMH - PubMed Abstract:
Tentoxin, a natural cyclic tetrapeptide produced by phytopathogenic fungi from the Alternaria species affects the catalytic function of the chloroplast F(1)-ATPase in certain sensitive species of plants. In this study, we show that the uncompetitive inhibitor tentoxin binds to the alphabeta-interface of the chloroplast F(1)-ATPase in a cleft localized at betaAsp-83. Most of the binding site is located on the noncatalytic alpha-subunit. The crystal structure of the tentoxin-inhibited CF(1)-complex suggests that the inhibitor is hydrogen bonded to Asp-83 in the catalytic beta-subunit but forms hydrophobic contacts with residues Ile-63, Leu-65, Val-75, Tyr-237, Leu-238, and Met-274 in the adjacent alpha-subunit. Except for minor changes around the tentoxin-binding site, the structure of the chloroplast alpha(3)beta(3)-core complex is the same as that determined with the native chloroplast ATPase. Tentoxin seems to act by inhibiting inter-subunit contacts at the alphabeta-interface and by blocking the interconversion of binding sites in the catalytic mechanism.
- Heinrich-Heine-Universität, Biochemie der Pflanzen, Universitätsstrasse 1, D-40225 Duesseldorf, Germany. georg.groth@uni-duesseldorf.de
Organizational Affiliation: