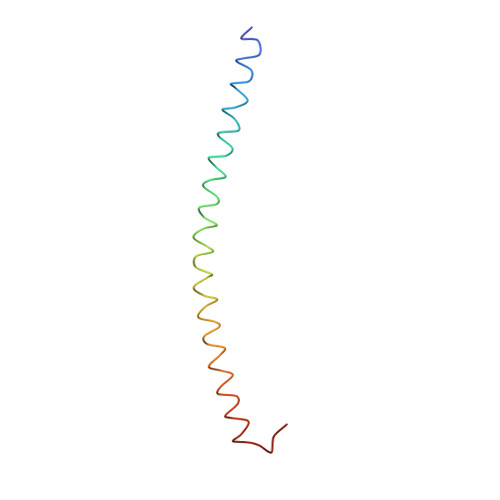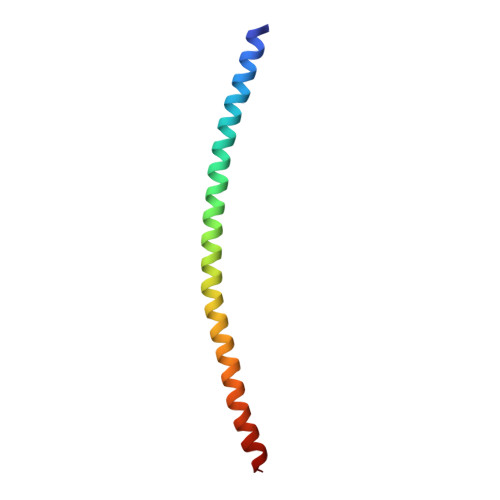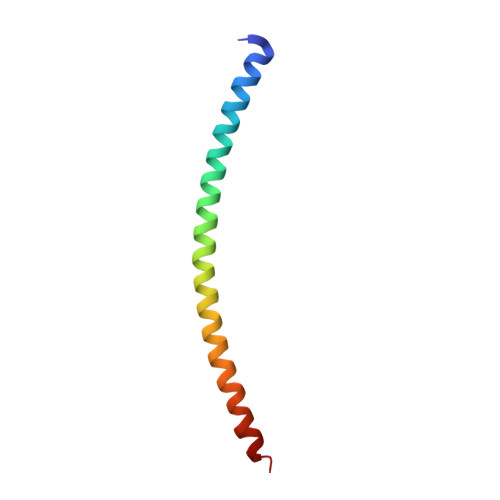Three-dimensional structure of the complexin/SNARE complex.
Chen, X., Tomchick, D.R., Kovrigin, E., Arac, D., Machius, M., Sudhof, T.C., Rizo, J.(2002) Neuron 33: 397-409
- PubMed: 11832227
- DOI: https://doi.org/10.1016/s0896-6273(02)00583-4
- Primary Citation of Related Structures:
1KIL - PubMed Abstract:
During neurotransmitter release, the neuronal SNARE proteins synaptobrevin/VAMP, syntaxin, and SNAP-25 form a four-helix bundle, the SNARE complex, that pulls the synaptic vesicle and plasma membranes together possibly causing membrane fusion. Complexin binds tightly to the SNARE complex and is essential for efficient Ca(2+)-evoked neurotransmitter release. A combined X-ray and TROSY-based NMR study now reveals the atomic structure of the complexin/SNARE complex. Complexin binds in an antiparallel alpha-helical conformation to the groove between the synaptobrevin and syntaxin helices. This interaction stabilizes the interface between these two helices, which bears the repulsive forces between the apposed membranes. These results suggest that complexin stabilizes the fully assembled SNARE complex as a key step that enables the exquisitely high speed of Ca(2+)-evoked neurotransmitter release.
- Department of Biochemistry, University of Texas Southwestern Medical Center, Dallas, TX 75390, USA.
Organizational Affiliation: