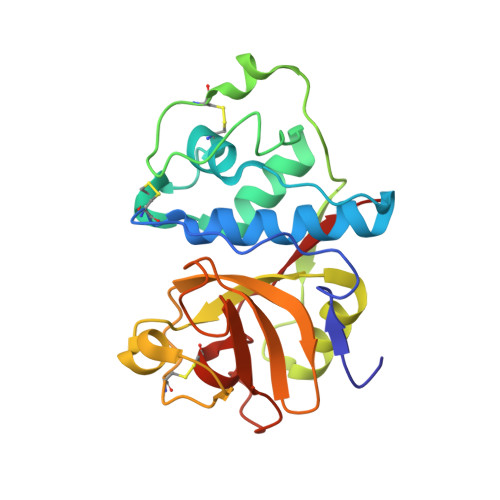
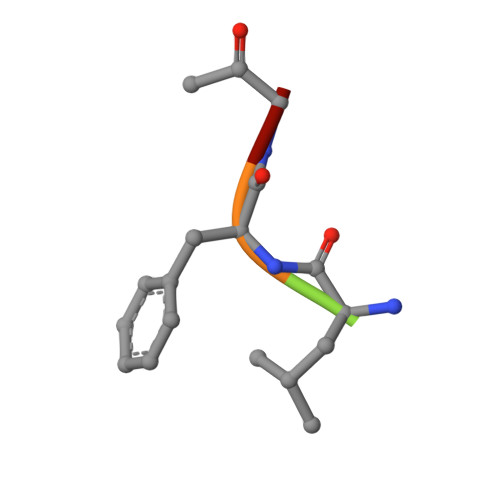
Two polymorphs of a covalent complex between papain and a diazomethylketone inhibitor
Janowski, R., Kozak, M., Jankowska, E., Grzonka, Z., Jaskolski, M.(2004) J Pept Res 64: 141-150
- PubMed: 15357669
- DOI: https://doi.org/10.1111/j.1399-3011.2004.00181.x
- Primary Citation of Related Structures:
1KHP, 1KHQ - PubMed Abstract:
The three-dimensional structure of two polymorphs of a ZLFG-CH2-papain covalent complex has been determined by X-ray crystallography. The structures indicate that: (i) the methylene carbon atom of the inhibitor is covalently bound to the Sgamma atom of Cys25 of papain; (ii) the hydrophobic S2 pocket formed by Pro68, Val133, Val157, and Asp158 is occupied by the inhibitor's phenylalanyl P2 side chain; (iii) extensive hydrogen bonding and hydrophobic interactions are responsible for the interaction of the inhibitor with the enzyme. Comparison with similar structures suggests that in covalent complexes preservation of main chain-main chain interactions between the enzyme and the inhibitor may have higher priority than the P-S interactions.
- Department of Crystallography, Faculty of Chemistry, A. Mickiewicz University, Grunwaldzka 6, 60-780 Poznań, Poland.
Organizational Affiliation: