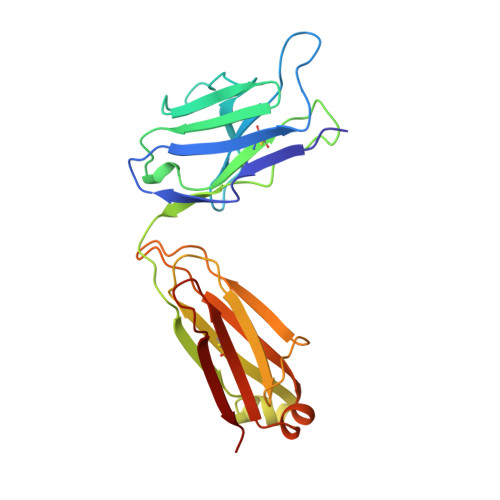
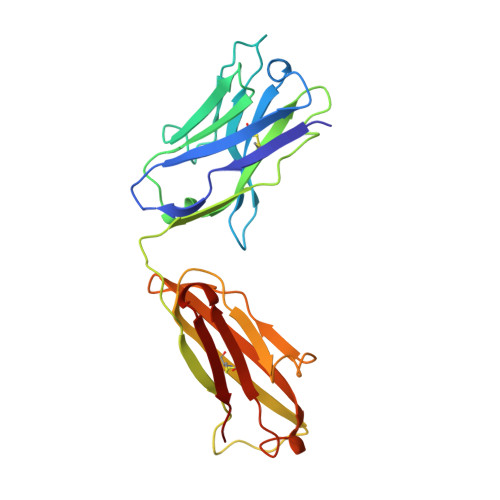
Insights into antibody catalysis: structure of an oxygenation catalyst at 1.9-angstrom resolution.
Hsieh-Wilson, L.C., Schultz, P.G., Stevens, R.C.(1996) Proc Natl Acad Sci U S A 93: 5363-5367
- PubMed: 8643580
- DOI: https://doi.org/10.1073/pnas.93.11.5363
- Primary Citation of Related Structures:
1KEL, 1KEM - PubMed Abstract:
The x-ray crystal structures of the sulfide oxidase antibody 28B4 and of antibody 28B4 complexed with hapten have been solved at 2.2-angstrom and 1.9-angstrom resolution, respectively. To our knowledge, these structures are the highest resolution catalytic antibody structures to date and provide insight into the molecular mechanism of this antibody-catalyzed monooxygenation reaction. Specifically, the data suggest that entropic restriction plays a fundamental role in catalysis through the precise alignment of the thioether substrate and oxidant. The antibody active site also stabilizes developing charge on both sulfur and periodate in the transition state via cation-pi and electrostatic interactions, respectively. In addition to demonstrating that the active site of antibody 28B4 does indeed reflect the mechanistic information programmed in the aminophosphonic acid hapten, these high-resolution structures provide a basis for enhancing turnover rates through mutagenesis and improved hapten design.
- Department of Chemistry, University of California, Berkeley, 94720, USA.
Organizational Affiliation: