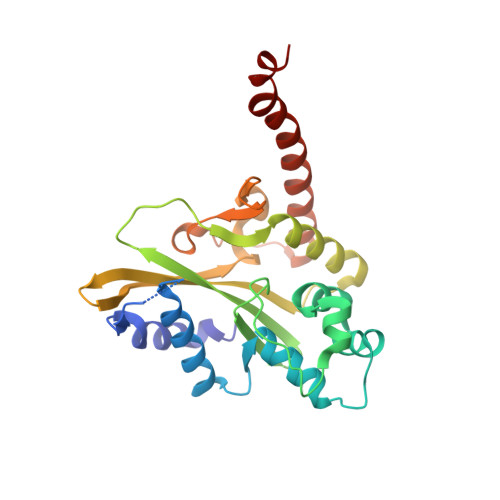
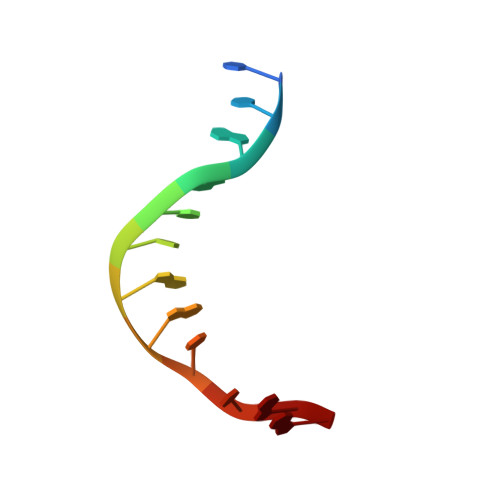
Sequence selectivity and degeneracy of a restriction endonuclease mediated by DNA intercalation.
Horton, N.C., Dorner, L.F., Perona, J.J.(2002) Nat Struct Biol 9: 42-47
- PubMed: 11742344
- DOI: https://doi.org/10.1038/nsb741
- Primary Citation of Related Structures:
1KC6 - PubMed Abstract:
The crystal structure of the HincII restriction endonuclease-DNA complex shows that degenerate specificity for blunt-ended cleavage at GTPyPuAC sequences arises from indirect readout of conformational preferences at the center pyrimidine-purine step. Protein-induced distortion of the DNA is accomplished by intercalation of glutamine side chains into the major groove on either side of the recognition site, generating bending by either tilt or roll at three distinct loci. The intercalated side chains propagate a concerted shift of all six target-site base pairs toward the minor groove, producing an unusual cross-strand purine stacking at the center pyrimidine-purine step. Comparison of the HincII and EcoRV cocrystal structures suggests that sequence-dependent differences in base-stacking free energies are a crucial underlying factor mediating protein recognition by indirect readout.
- Department of Chemistry and Biochemistry, and Interdepartmental Program in Biomolecular Science and Engineering, University of California at Santa Barbara, Santa Barbara, California 93106-9510, USA.
Organizational Affiliation: