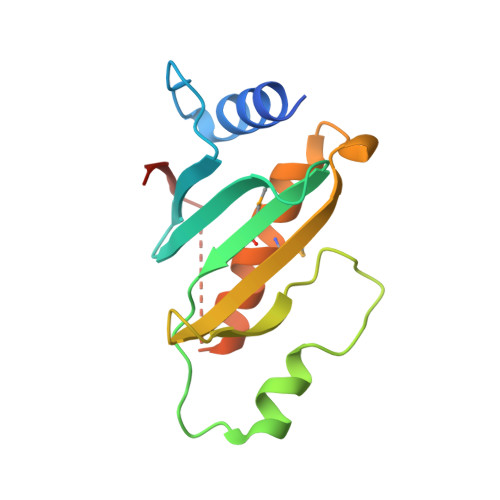
Three-dimensional structure of the type III secretion chaperone SycE from Yersinia pestis.
Evdokimov, A.G., Tropea, J.E., Routzahn, K.M., Waugh, D.S.(2002) Acta Crystallogr D Biol Crystallogr 58: 398-406
- PubMed: 11856824
- DOI: https://doi.org/10.1107/s090744490200015x
- Primary Citation of Related Structures:
1K6Z - PubMed Abstract:
Many bacterial pathogens utilize a type III (contact-dependent) secretion system to inject cytotoxic effector proteins directly into host cells. This ingenious mechanism, designed for both bacterial offense and defense, has been studied most extensively in Yersinia spp. To be exported efficiently, at least three of the effectors (YopE, YopH and YopT) and several other proteins that transit the type III secretion pathway in Yersinia (YopN, YopD and YopB) must first form transient complexes with cognate-specific Yop chaperone (Syc) proteins. The cytotoxic effector YopE, a selective activator of mammalian Rho-family GTPases, associates with SycE. Here, the structure of Y. pestis SycE at 1.95A resolution is reported. SycE possesses a novel fold with an unusual dimerization motif and an intriguing basic cavity located on the dyad axis of the dimer that may participate in its interaction with YopE.
- Macromolecular Crystallography Laboratory, Center for Cancer Research, National Cancer Institute at Frederick, PO Box B, Frederick, MD 21702-1201, USA.
Organizational Affiliation: