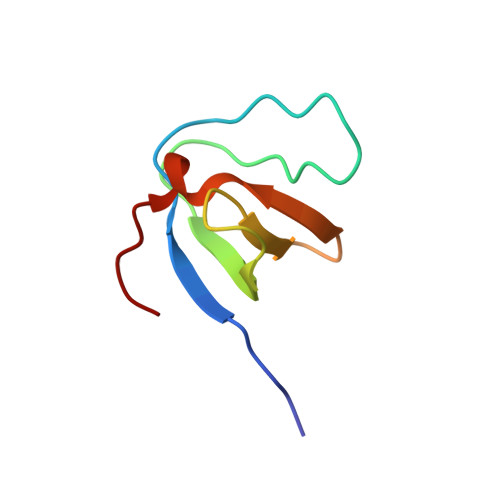
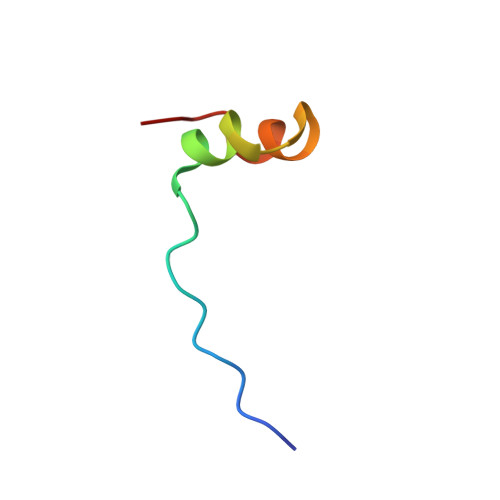
Diverse recognition of non-PxxP peptide ligands by the SH3 domains from p67(phox), Grb2 and Pex13p.
Kami, K., Takeya, R., Sumimoto, H., Kohda, D.(2002) EMBO J 21: 4268-4276
- PubMed: 12169629
- DOI: https://doi.org/10.1093/emboj/cdf428
- Primary Citation of Related Structures:
1K4U - PubMed Abstract:
The basic function of the Src homology 3 (SH3) domain is considered to be binding to proline-rich sequences containing a PxxP motif. Recently, many SH3 domains, including those from Grb2 and Pex13p, were reported to bind sequences lacking a PxxP motif. We report here that the 22 residue peptide lacking a PxxP motif, derived from p47(phox), binds to the C-terminal SH3 domain from p67(phox). We applied the NMR cross-saturation method to locate the interaction sites for the non-PxxP peptides on their cognate SH3 domains from p67(phox), Grb2 and Pex13p. The binding site of the Grb2 SH3 partially overlapped the conventional PxxP-binding site, whereas those of p67(phox) and Pex13p SH3s are located in different surface regions. The non-PxxP peptide from p47(phox) binds to the p67(phox) SH3 more tightly when it extends to the N-terminus to include a typical PxxP motif, which enabled the structure determination of the complex, to reveal that the non-PxxP peptide segment interacted with the p67(phox) SH3 in a compact helix-turn-helix structure (PDB entry 1K4U).
- Department of Structural Biology, Biomolecular Engineering Research Institute, 6-2-3 Furuedai, Suita, Osaka 565-0874, Japan.
Organizational Affiliation: