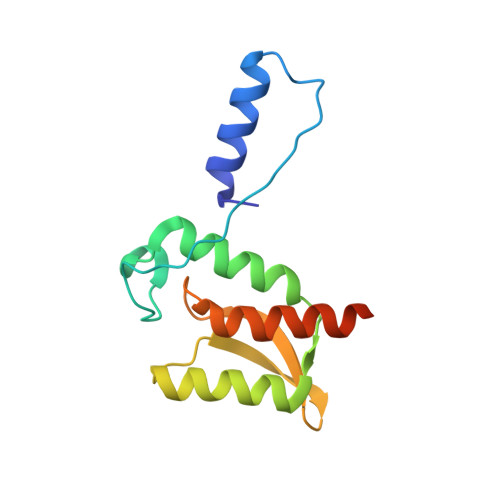
Structure of the type III secretion and substrate-binding domain of Yersinia YopH phosphatase.
Smith, C.L., Khandelwal, P., Keliikuli, K., Zuiderweg, E.R., Saper, M.A.(2001) Mol Microbiol 42: 967-979
- PubMed: 11737640
- DOI: https://doi.org/10.1046/j.0950-382x.2001.02711.x
- Primary Citation of Related Structures:
1K46 - PubMed Abstract:
Pathogenic strains of Yersinia deploy a type III secretion system to inject the potent tyrosine phosphatase YopH into host cells, where it dephosphorylates focal adhesion-associated substrates. The amino-terminal, non-catalytic domain of YopH is bifunctional; it is essential for the secretion and binding of the specific chaperone SycH, but also targets the catalytic domain to substrates in the infected cell. We describe the 2.2 A resolution crystal structure of residues 1-129 of YopH from Yersinia pseudotuberculosis. The amino-terminal alpha-helix (2-17), comprising the secretion signal, and beta-strand (24-28) of one molecule exchange with another molecule to form a domain-swapped dimer. Nuclear magnetic resonance (NMR) and gel filtration experiments demonstrated that YopH(1-129) could exist as a monomer and/or a dimer in solution. The topology of the dimer and the dynamics of a monomeric form in solution observed by NMR imply that YopH has the propensity to unfold partially. The dimer is probably not important physiologically, but may mimic how SycH binds to the exposed non-polar surfaces of a partially unfolded YopH. Phosphopeptide-induced perturbations in NMR chemical shifts define a substrate-binding surface on YopH(1-129) that includes residues previously shown by mutagenesis to be essential for YopH function.
- Department of Biological Chemistry, University of Michigan, 930 N. University Ave., Ann Arbor, MI 48109-1055, USA.
Organizational Affiliation: