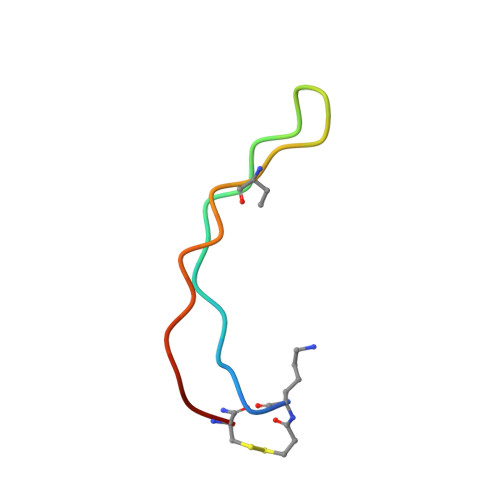
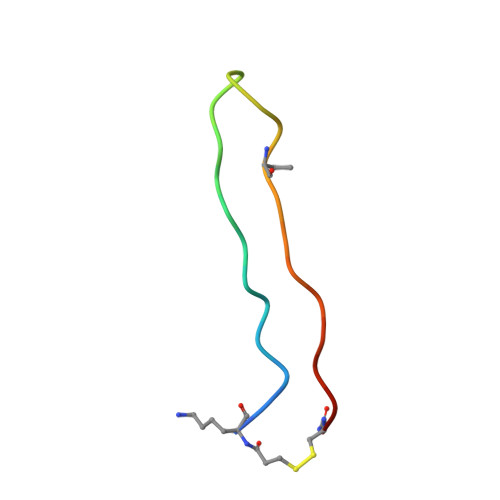
BetaCore, a designed water soluble four-stranded antiparallel beta-sheet protein.
Carulla, N., Woodward, C., Barany, G.(2002) Protein Sci 11: 1539-1551
- PubMed: 12021452
- DOI: https://doi.org/10.1110/ps.4440102
- Primary Citation of Related Structures:
1K09 - PubMed Abstract:
BetaCore is a designed approximately 50-residue protein in which two BPTI-derived core modules, CM I and CM II, are connected by a 22-atom cross-link. At low temperature and pH 3, homo- and heteronuclear NMR data report a dominant folded ('f') conformation with well-dispersed chemical shifts, i, i+1 periodicity, numerous long-range NOEs, and slowed amide hydrogen isotope exchange patterns that is a four-stranded antiparallel beta-sheet with nonsymmetrical and specific association of CM I and CM II. BetaCore 'f' conformations undergo reversible, global, moderately cooperative, non-two-state thermal transitions to an equilibrium ensemble of unfolded 'u' conformations. There is a significant energy barrier between 'f' and 'u' conformations. This is the first designed four-stranded antiparallel beta-sheet that folds in water.
- Department of Chemistry, University of Minnesota, Minneapolis, Minnesota 55455, USA.
Organizational Affiliation: