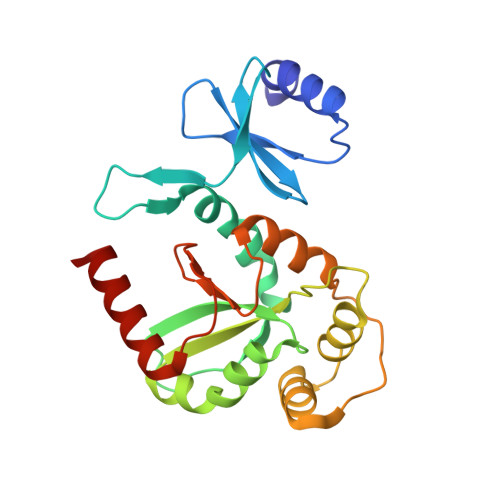
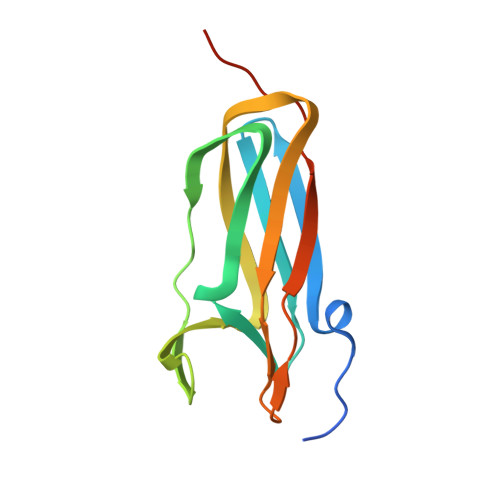
The Disulfide Bond Isomerase DsbC is Activated by an Immunoglobulin-fold Thiol Oxidoreductase: Crystal structure of the DsbC-DsbDalpha complex.
Haebel, P.W., Goldstone, D., Katzen, F., Beckwith, J., Metcalf, P.(2002) EMBO J 21: 4774-4784
- PubMed: 12234918
- DOI: https://doi.org/10.1093/emboj/cdf489
- Primary Citation of Related Structures:
1JPE, 1JZD, 1JZO - PubMed Abstract:
The Escherichia coli disulfide bond isomerase DsbC rearranges incorrect disulfide bonds during oxidative protein folding. It is specifically activated by the periplasmic N-terminal domain (DsbDalpha) of the transmembrane electron transporter DsbD. An intermediate of the electron transport reaction was trapped, yielding a covalent DsbC-DsbDalpha complex. The 2.3 A crystal structure of the complex shows for the first time the specific interactions between two thiol oxidoreductases. DsbDalpha is a novel thiol oxidoreductase with the active site cysteines embedded in an immunoglobulin fold. It binds into the central cleft of the V-shaped DsbC dimer, which assumes a closed conformation on complex formation. Comparison of the complex with oxidized DsbDalpha reveals major conformational changes in a cap structure that regulates the accessibility of the DsbDalpha active site. Our results explain how DsbC is selectively activated by DsbD using electrons derived from the cytoplasm.
- School of Biological Sciences, University of Auckland, Private Bag 92019, Auckland, New Zealandand.
Organizational Affiliation: