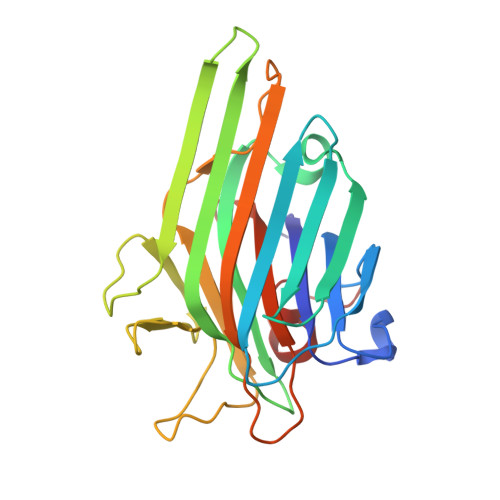
Structural and Functional Consequences of Peptide-carbohydrate Mimicry. Crystal Structure of a Carbohydrate-mimicking Peptide Bound to Concanavalin A.
Jain, D., Kaur, K.J., Sundaravadivel, B., Salunke, D.M.(2000) J Biological Chem 275: 16098-16102
- PubMed: 10821862
- DOI: https://doi.org/10.1074/jbc.275.21.16098
- Primary Citation of Related Structures:
1JYI - PubMed Abstract:
The functional consequences of peptide-carbohydrate mimicry were analyzed on the basis of the crystal structure of concanavalin A (ConA) in complex with a carbohydrate-mimicking peptide, DVFYPYPYASGS. The peptide binds to the non-crystallographically related monomers of two independent dimers of ConA in two different modes, in slightly different conformations, demonstrating structural adaptability in ConA-peptide recognition. In one mode, the peptide has maximum interactions with ConA, and in the other, it shows relatively fewer contacts within this site but significant contacts with the symmetry-related subunit. Neither of the peptide binding sites overlaps with the structurally characterized mannose and trimannose binding sites on ConA. Despite this, the functional mimicry between the peptide and carbohydrate ligands was evident. The peptide-inhibited ConA induced T cell proliferation in a dose-dependent manner. The effect of the designed analogs of the peptide on ConA-induced T cell proliferation and their recognition by the antibody response against alpha-d-mannopyranoside indicate a role for aromatic residues in functional mimicry. Although the functional mimicry was observed between the peptide and carbohydrate moieties, the crystal structure of the ConA-peptide complex revealed that the two peptide binding sites are independent of the methyl alpha-d-mannopyranoside binding site.
- Structural Biology Unit, National Institute of Immunology, Aruna Asaf Ali Marg, New Delhi 110 067, India.
Organizational Affiliation: