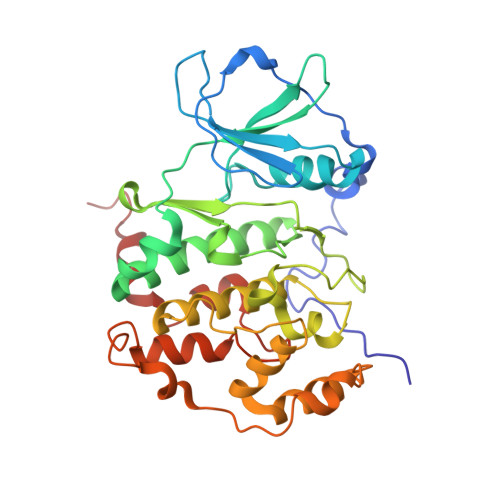
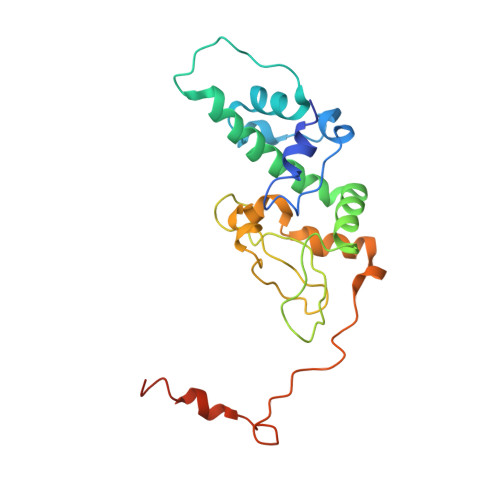
Crystal structure of human protein kinase CK2: insights into basic properties of the CK2 holoenzyme.
Niefind, K., Guerra, B., Ermakowa, I., Issinger, O.G.(2001) EMBO J 20: 5320-5331
- PubMed: 11574463
- DOI: https://doi.org/10.1093/emboj/20.19.5320
- Primary Citation of Related Structures:
1JWH - PubMed Abstract:
The crystal structure of a fully active form of human protein kinase CK2 (casein kinase 2) consisting of two C-terminally truncated catalytic and two regulatory subunits has been determined at 3.1 A resolution. In the CK2 complex the regulatory subunits form a stable dimer linking the two catalytic subunits, which make no direct contact with one another. Each catalytic subunit interacts with both regulatory chains, predominantly via an extended C-terminal tail of the regulatory subunit. The CK2 structure is consistent with its constitutive activity and with a flexible role of the regulatory subunit as a docking partner for various protein kinases. Furthermore it shows an inter-domain mobility in the catalytic subunit known to be functionally important in protein kinases and detected here for the first time directly within one crystal structure.
- Universität zu Köln, Institut für Biochemie, Zülpicher Strasse 47, D-50674 Köln, Germany. Karsten.Niefind@uni-koeln.de
Organizational Affiliation: