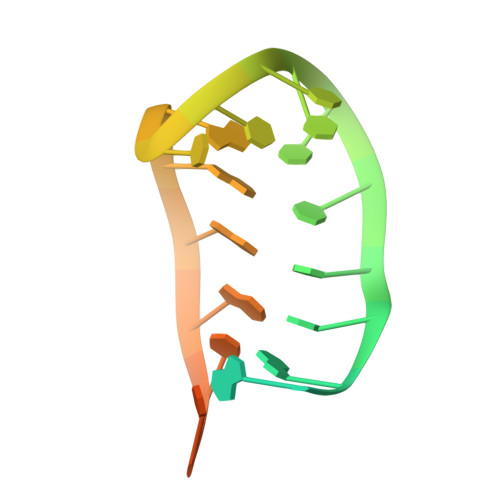
NMR structure and dynamics of the RNA-binding site for the histone mRNA stem-loop binding protein.
DeJong, E.S., Marzluff, W.F., Nikonowicz, E.P.(2002) RNA 8: 83-96
- PubMed: 11871662
- DOI: https://doi.org/10.1017/s1355838202013869
- Primary Citation of Related Structures:
1JU7, 1JWC - PubMed Abstract:
The 3' end of replication-dependent histone mRNAs terminate in a conserved sequence containing a stem-loop. This 26-nt sequence is the binding site for a protein, stem-loop binding protein (SLBP), that is involved in multiple aspects of histone mRNA metabolism and regulation. We have determined the structure of the 26-nt sequence by multidimensional NMR spectroscopy. There is a 16-nt stem-loop motif, with a conserved 6-bp stem and a 4-nt loop. The loop is closed by a conserved U.A base pair that terminates the canonical A-form stem. The pyrimidine-rich 4-nt loop, UUUC, is well organized with the three uridines stacking on the helix, and the fourth base extending across the major groove into the solvent. The flanking nucleotides at the base of the hairpin stem do not assume a unique conformation, despite the fact that the 5' flanking nucleotides are a critical component of the SLBP binding site.
- Department of Biochemistry and Cell Biology, Rice University, Houston, Texas 77251-1892, USA.
Organizational Affiliation: