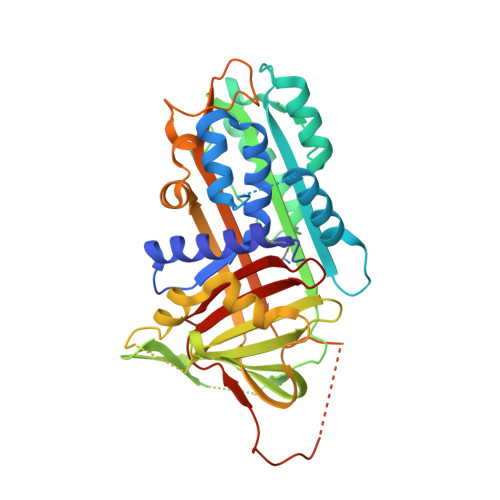
CRYSTAL STRUCTURE OF THE COMPLEX OF PLASMINOGEN ACTIVATOR INHIBITOR 2 WITH A PEPTIDE MIMICKING THE REACTIVE CENTER LOOP
JANKOVA, L., HARROP, S.J., SAUNDERS, D.N., ANDREWS, J.L., BERTRAM, K.C., GOULD, A.R., BAKER, M.S., CURMI, P.M.G.(2001) J Biological Chem 276: 43374-43382
- PubMed: 11546761
- DOI: https://doi.org/10.1074/jbc.M103021200
- Primary Citation of Related Structures:
1JRR - PubMed Abstract:
The structure of the serpin, plasminogen activator inhibitor type-2 (PAI-2), in a complex with a peptide mimicking its reactive center loop (RCL) has been determined at 1.6-A resolution. The structure shows the relaxed state serpin structure with a prominent six-stranded beta-sheet. Clear electron density is seen for all residues in the peptide. The P1 residue of the peptide binds to a well defined pocket at the base of PAI-2 that may be important in determining the specificity of protease inhibition. The stressed-to-relaxed state (S --> R) transition in PAI-2 can be modeled as the relative motion between a quasirigid core domain and a smaller segment comprising helix hF and beta-strands s1A, s2A, and s3A. A comparison of the Ramachandran plots of the stressed and relaxed state PAI-2 structures reveals the location of several hinge regions connecting these two domains. The hinge regions cluster in three locations on the structure, ensuring a cooperative S --> R transition. We hypothesize that the hinge formed by the conserved Gly(206) on beta-strand s3A in the breach region of PAI-2 effects the S --> R transition by altering its backbone torsion angles. This torsional change is due to the binding of the P14 threonine of the RCL to the open breach region of PAI-2.
- Initiative in Biomolecular Structure, School of Physics, University of New South Wales, Sydney New South Wales 2052, Australia.
Organizational Affiliation: