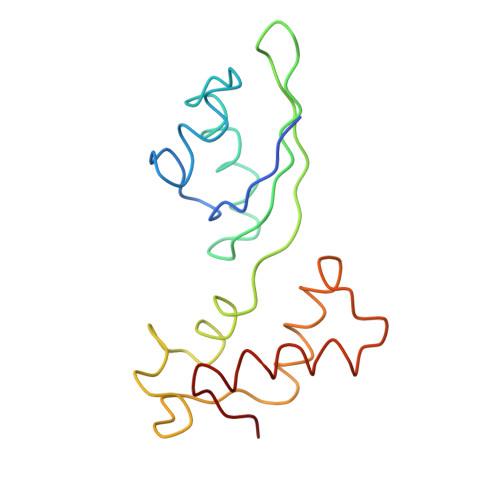
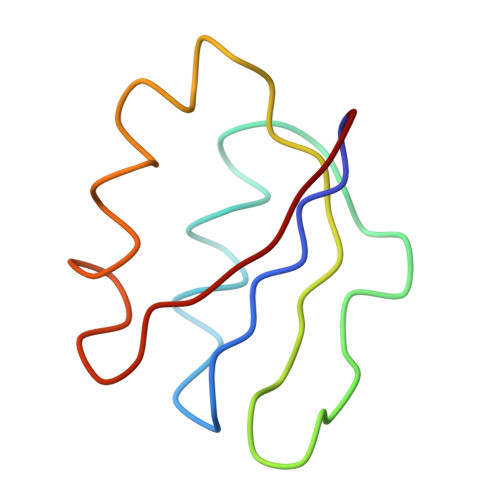
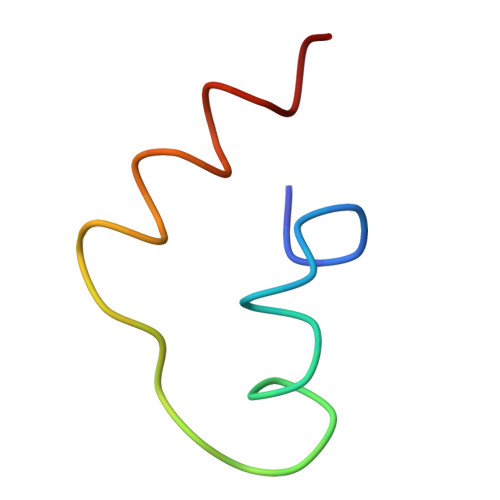Localization of L11 protein on the ribosome and elucidation of its involvement in EF-G-dependent translocation.
Agrawal, R.K., Linde, J., Sengupta, J., Nierhaus, K.H., Frank, J.(2001) J Mol Biology 311: 777-787
- PubMed: 11518530
- DOI: https://doi.org/10.1006/jmbi.2001.4907
- Primary Citation of Related Structures:
1JQM, 1JQS, 1JQT - PubMed Abstract:
L11 protein is located at the base of the L7/L12 stalk of the 50 S subunit of the Escherichia coli ribosome. Because of the flexible nature of the region, recent X-ray crystallographic studies of the 50 S subunit failed to locate the N-terminal domain of the protein. We have determined the position of the complete L11 protein by comparing a three-dimensional cryo-EM reconstruction of the 70 S ribosome, isolated from a mutant lacking ribosomal protein L11, with the three-dimensional map of the wild-type ribosome. Fitting of the X-ray coordinates of L11-23 S RNA complex and EF-G into the cryo-EM maps combined with molecular modeling, reveals that, following EF-G-dependent GTP hydrolysis, domain V of EF-G intrudes into the cleft between the 23 S ribosomal RNA and the N-terminal domain of L11 (where the antibiotic thiostrepton binds), causing the N-terminal domain to move and thereby inducing the formation of the arc-like connection with the G' domain of EF-G. The results provide a new insight into the mechanism of EF-G-dependent translocation.
- Wadsworth Center, Empire State Plaza, Albany, NY 12201-0509, USA. agrawal@wadsworth.org
Organizational Affiliation: