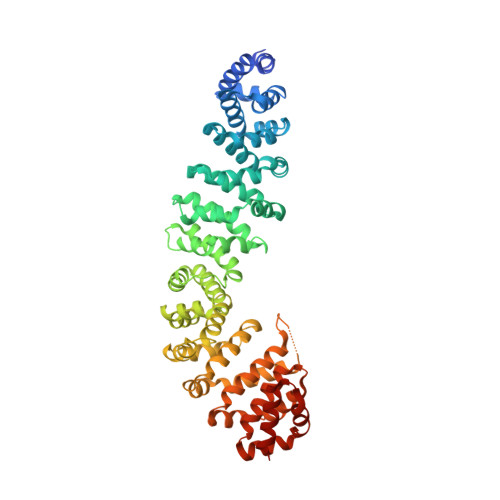
Molecular mechanisms of beta-catenin recognition by adenomatous polyposis coli revealed by the structure of an APC-beta-catenin complex.
Eklof Spink, K., Fridman, S.G., Weis, W.I.(2001) EMBO J 20: 6203-6212
- PubMed: 11707392
- DOI: https://doi.org/10.1093/emboj/20.22.6203
- Primary Citation of Related Structures:
1JPP - PubMed Abstract:
The adenomatous polyposis coli (APC) tumor suppressor protein plays a critical role in regulating cellular levels of the oncogene product beta-catenin. APC binds to beta-catenin through a series of homologous 15 and 20 amino acid repeats. We have determined the crystal structure of a 15 amino acid beta-catenin binding repeat from APC bound to the armadillo repeat region of beta-catenin. Although it lacks significant sequence homology, the N-terminal half of the repeat binds in a manner similar to portions of E-cadherin and XTcf3, but the remaining interactions are unique to APC. We discuss the implications of this new structure for the design of therapeutics, and present evidence from structural, biochemical and sequence data, which suggest that the 20 amino acid repeats can adopt two modes of binding to beta-catenin.
- Departments of Structural Biology and of Molecular and Cellular Physiology, Stanford University School of Medicine, 299 Campus Dr. West Stanford, CA 94305, USA.
Organizational Affiliation: