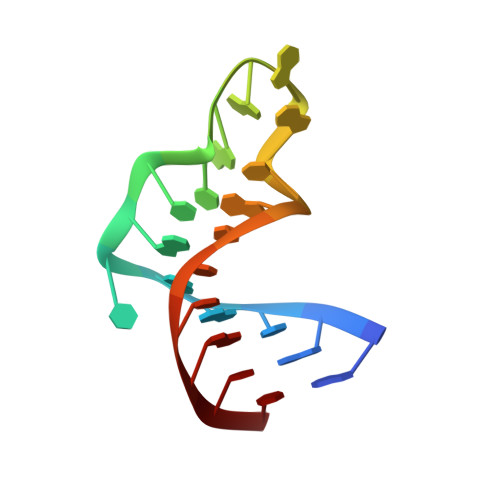
Structure of the UGAGAU hexaloop that braces Bacillus RNase P for action.
Leeper, T.C., Martin, M.B., Kim, H., Cox, S., Semenchenko, V., Schmidt, F.J., Van Doren, S.R.(2002) Nat Struct Biol 9: 397-403
- PubMed: 11927952
- DOI: https://doi.org/10.1038/nsb775
- Primary Citation of Related Structures:
1JOX, 1JP0 - PubMed Abstract:
Long-range interactions involving the P5.1 hairpin of Bacillus RNase P RNA are thought to form a structural truss to support RNA folding and activity. We determined the structure of this element by NMR and refined the structure using residual dipolar couplings from a sample weakly oriented in a dilute liquid crystalline mixture of polyethylene glycol and hexanol. Dipolar coupling refinement improved the global precision of the structure from 1.5 to 1.2 A (to the mean), revised the bend angle between segments of the P5.1 stem and corroborated the structure of the loop region. The UGAGAU hexaloop of P5.1 contains two stacks of bases on opposite sides of the loop, distinguishing it from GNRA tetraloops. The unusual conformation of the juxtaposed uracil residues within the hexaloop may explain their requirement in transactivation assays.
- Department of Biochemistry, 117 Schweitzer Hall, University of Missouri, Columbia, Missouri 65211, USA.
Organizational Affiliation: